if(!require(pacman))
install.packages("pacman")
pacman::p_load(
C50, # C5.0 Decision Trees and Rule-Based Models
caret, # Classification and Regression Training
e1071, # Misc Functions of the Department of Statistics (e1071), TU Wien
keras, # R Interface to 'Keras'
kernlab, # Kernel-Based Machine Learning Lab
lattice, # Trellis Graphics for R
MASS, # Support Functions and Datasets for Venables and Ripley's MASS
mlbench, # Machine Learning Benchmark Problems
nnet, # Feedforward Neural Networks and Multinomial Log-Linear Models
palmerpenguins, # Palmer Archipelago (Antarctica) Penguin Data
party, # A Laboratory for Recursive Partytioning
partykit, # A Toolkit for Recursive Partytioning
randomForest, # Breiman and Cutler's Random Forests for Classification and Regression
rpart, # Recursive partitioning models
RWeka, # R/Weka Interface
scales, # Scale Functions for Visualization
tidymodels, # Tidy machine learning framework
tidyverse, # Tidy data wrangling and visualization
xgboost # Extreme Gradient Boosting
)Classification: Alternative Techniques
Install packages
Install the packages used in this chapter:
Show fewer digits
options(digits=3)Introduction
Many different classification algorithms have been proposed in the literature. In this chapter, we will apply some of the more popular methods.
Training and Test Data
We will use the Zoo dataset which is included in the R package mlbench (you may have to install it). The Zoo dataset containing 17 (mostly logical) variables on different 101 animals as a data frame with 17 columns (hair, feathers, eggs, milk, airborne, aquatic, predator, toothed, backbone, breathes, venomous, fins, legs, tail, domestic, catsize, type). We convert the data frame into a tidyverse tibble (optional).
data(Zoo, package="mlbench")
Zoo <- as.data.frame(Zoo)
Zoo |> glimpse()Rows: 101
Columns: 17
$ hair <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE…
$ feathers <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE…
$ eggs <lgl> FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, F…
$ milk <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE…
$ airborne <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE…
$ aquatic <lgl> FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, F…
$ predator <lgl> TRUE, FALSE, TRUE, TRUE, TRUE, FALSE, FALSE, FALSE, TRUE, FAL…
$ toothed <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, T…
$ backbone <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, T…
$ breathes <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE…
$ venomous <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE…
$ fins <lgl> FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, F…
$ legs <int> 4, 4, 0, 4, 4, 4, 4, 0, 0, 4, 4, 2, 0, 0, 4, 6, 2, 4, 0, 0, 2…
$ tail <lgl> FALSE, TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, TRUE, FALSE…
$ domestic <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, FALSE, …
$ catsize <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, FALS…
$ type <fct> mammal, mammal, fish, mammal, mammal, mammal, mammal, fish, f…We will use the package caret to make preparing training sets and building classification (and regression) models easier. A great cheat sheet can be found here.
Multi-core support can be used for cross-validation. Note: It is commented out here because it does not work with rJava used in RWeka below.
##library(doMC, quietly = TRUE)
##registerDoMC(cores = 4)
##getDoParWorkers()Test data is not used in the model building process and needs to be set aside purely for testing the model after it is completely built. Here I use 80% for training.
set.seed(123) # for reproducibility
inTrain <- createDataPartition(y = Zoo$type, p = .8)[[1]]
Zoo_train <- dplyr::slice(Zoo, inTrain)
Zoo_test <- dplyr::slice(Zoo, -inTrain)Fitting Different Classification Models to the Training Data
Create a fixed sampling scheme (10-folds) so we can compare the fitted models later.
train_index <- createFolds(Zoo_train$type, k = 10)The fixed folds are used in train() with the argument trControl = trainControl(method = "cv", indexOut = train_index)). If you don’t need fixed folds, then remove indexOut = train_index in the code below.
For help with building models in caret see: ? train
Note: Be careful if you have many NA values in your data. train() and cross-validation many fail in some cases. If that is the case then you can remove features (columns) which have many NAs, omit NAs using na.omit() or use imputation to replace them with reasonable values (e.g., by the feature mean or via kNN). Highly imbalanced datasets are also problematic since there is a chance that a fold does not contain examples of each class leading to a hard to understand error message.
Conditional Inference Tree (Decision Tree)
ctreeFit <- Zoo_train |> train(type ~ .,
method = "ctree",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index))
ctreeFitConditional Inference Tree
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 76, 72, 73, 76, 75, 75, ...
Resampling results across tuning parameters:
mincriterion Accuracy Kappa
0.010 0.827 0.772
0.255 0.827 0.772
0.500 0.827 0.772
0.745 0.827 0.772
0.990 0.827 0.772
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mincriterion = 0.99.plot(ctreeFit$finalModel)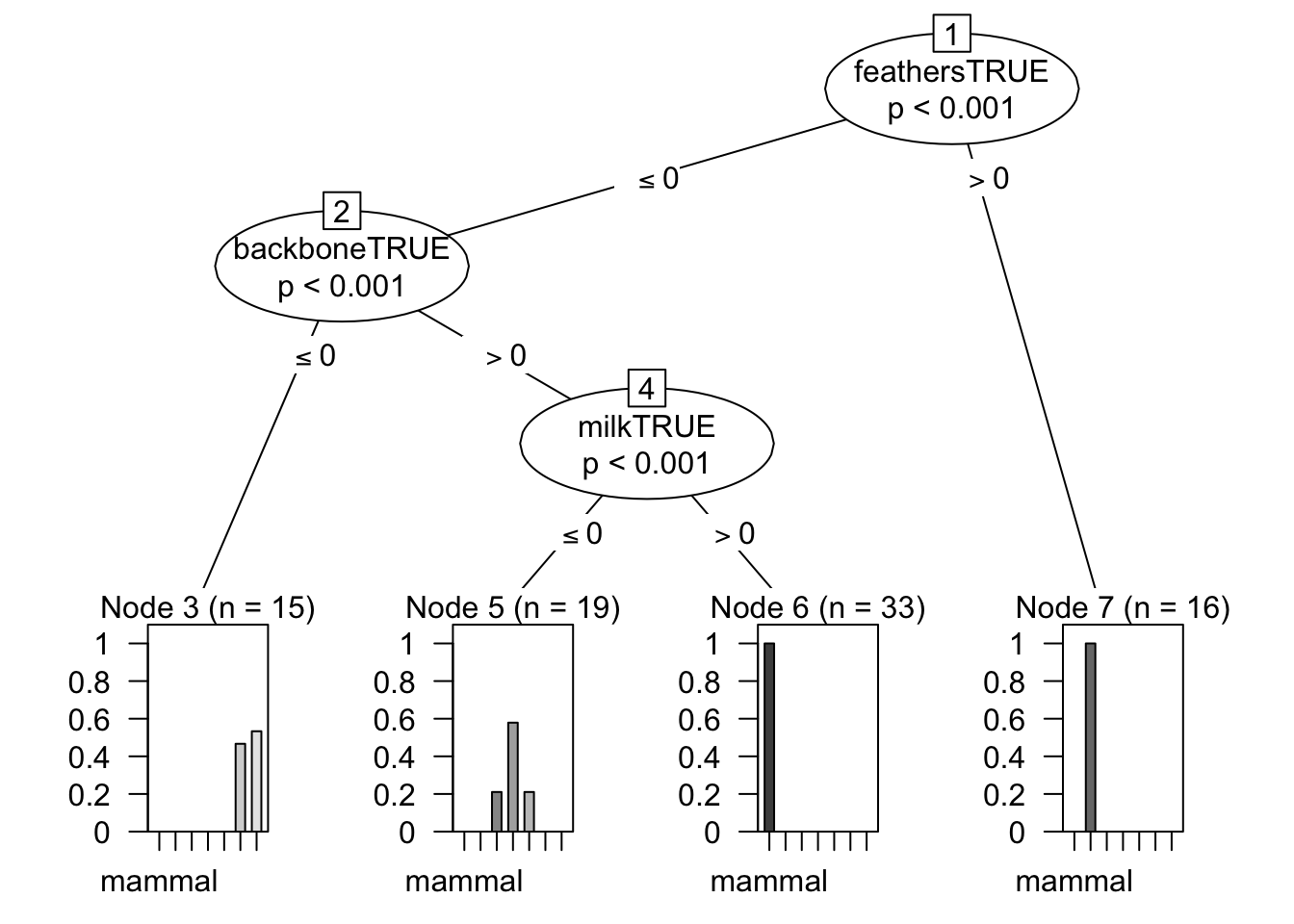
C 4.5 Decision Tree
C45Fit <- Zoo_train |> train(type ~ .,
method = "J48",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index))
C45FitC4.5-like Trees
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 76, 75, 73, 76, 74, 74, ...
Resampling results across tuning parameters:
C M Accuracy Kappa
0.010 1 0.975 0.967
0.010 2 0.965 0.954
0.010 3 0.953 0.940
0.010 4 0.959 0.948
0.010 5 0.970 0.962
0.133 1 1.000 1.000
0.133 2 0.976 0.968
0.133 3 0.965 0.954
0.133 4 0.959 0.948
0.133 5 0.970 0.962
0.255 1 1.000 1.000
0.255 2 0.976 0.968
0.255 3 0.965 0.954
0.255 4 0.959 0.948
0.255 5 0.970 0.962
0.378 1 1.000 1.000
0.378 2 0.976 0.968
0.378 3 0.965 0.954
0.378 4 0.959 0.948
0.378 5 0.970 0.962
0.500 1 1.000 1.000
0.500 2 0.976 0.968
0.500 3 0.965 0.954
0.500 4 0.959 0.948
0.500 5 0.970 0.962
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were C = 0.133 and M = 1.C45Fit$finalModelJ48 pruned tree
------------------
feathersTRUE <= 0
| milkTRUE <= 0
| | backboneTRUE <= 0
| | | predatorTRUE <= 0
| | | | legs <= 2: mollusc.et.al (1.0)
| | | | legs > 2: insect (6.0)
| | | predatorTRUE > 0: mollusc.et.al (8.0/1.0)
| | backboneTRUE > 0
| | | finsTRUE <= 0
| | | | aquaticTRUE <= 0: reptile (3.0)
| | | | aquaticTRUE > 0
| | | | | eggsTRUE <= 0: reptile (1.0)
| | | | | eggsTRUE > 0: amphibian (4.0)
| | | finsTRUE > 0: fish (11.0)
| milkTRUE > 0: mammal (33.0)
feathersTRUE > 0: bird (16.0)
Number of Leaves : 9
Size of the tree : 17K-Nearest Neighbors
Note: kNN uses Euclidean distance, so data should be standardized (scaled) first. Here legs are measured between 0 and 6 while all other variables are between 0 and 1. Scaling can be directly performed as preprocessing in train using the parameter preProcess = "scale".
knnFit <- Zoo_train |> train(type ~ .,
method = "knn",
data = _,
preProcess = "scale",
tuneLength = 5,
tuneGrid=data.frame(k = 1:10),
trControl = trainControl(method = "cv", indexOut = train_index))
knnFitk-Nearest Neighbors
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
Pre-processing: scaled (16)
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 77, 74, 75, 75, 74, 74, ...
Resampling results across tuning parameters:
k Accuracy Kappa
1 1.000 1.000
2 0.965 0.954
3 0.963 0.951
4 0.942 0.925
5 0.941 0.921
6 0.963 0.951
7 0.963 0.951
8 0.941 0.921
9 0.908 0.883
10 0.918 0.892
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was k = 1.knnFit$finalModel1-nearest neighbor model
Training set outcome distribution:
mammal bird reptile fish amphibian
33 16 4 11 4
insect mollusc.et.al
7 8 PART (Rule-based classifier)
rulesFit <- Zoo_train |> train(type ~ .,
method = "PART",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index))
rulesFitRule-Based Classifier
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 73, 74, 76, 76, 75, 74, ...
Resampling results across tuning parameters:
threshold pruned Accuracy Kappa
0.010 yes 0.979 0.973
0.010 no 0.979 0.973
0.133 yes 0.990 0.987
0.133 no 0.979 0.973
0.255 yes 0.990 0.987
0.255 no 0.979 0.973
0.378 yes 0.990 0.987
0.378 no 0.979 0.973
0.500 yes 0.990 0.987
0.500 no 0.979 0.973
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were threshold = 0.5 and pruned = yes.rulesFit$finalModelPART decision list
------------------
feathersTRUE <= 0 AND
milkTRUE > 0: mammal (33.0)
feathersTRUE > 0: bird (16.0)
backboneTRUE <= 0 AND
airborneTRUE <= 0 AND
predatorTRUE > 0: mollusc.et.al (7.0)
backboneTRUE > 0 AND
finsTRUE > 0: fish (11.0)
backboneTRUE <= 0: insect (8.0/1.0)
aquaticTRUE > 0: amphibian (5.0/1.0)
: reptile (3.0)
Number of Rules : 7Linear Support Vector Machines
svmFit <- Zoo_train |> train(type ~.,
method = "svmLinear",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index))
svmFitSupport Vector Machines with Linear Kernel
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 74, 74, 77, 75, 74, 77, ...
Resampling results:
Accuracy Kappa
1 1
Tuning parameter 'C' was held constant at a value of 1svmFit$finalModelSupport Vector Machine object of class "ksvm"
SV type: C-svc (classification)
parameter : cost C = 1
Linear (vanilla) kernel function.
Number of Support Vectors : 39
Objective Function Value : -0.143 -0.217 -0.15 -0.175 -0.0934 -0.0974 -0.292 -0.0835 -0.154 -0.0901 -0.112 -0.189 -0.593 -0.13 -0.179 -0.122 -0.0481 -0.0838 -0.125 -0.15 -0.501
Training error : 0 Random Forest
randomForestFit <- Zoo_train |> train(type ~ .,
method = "rf",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index))
randomForestFitRandom Forest
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 75, 76, 75, 76, 74, 73, ...
Resampling results across tuning parameters:
mtry Accuracy Kappa
2 1 1
5 1 1
9 1 1
12 1 1
16 1 1
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mtry = 2.randomForestFit$finalModel
Call:
randomForest(x = x, y = y, mtry = param$mtry)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 2
OOB estimate of error rate: 7.23%
Confusion matrix:
mammal bird reptile fish amphibian insect mollusc.et.al
mammal 33 0 0 0 0 0 0
bird 0 16 0 0 0 0 0
reptile 0 1 0 2 1 0 0
fish 0 0 0 11 0 0 0
amphibian 0 0 0 0 4 0 0
insect 0 0 0 0 0 7 0
mollusc.et.al 1 0 0 0 0 1 6
class.error
mammal 0.00
bird 0.00
reptile 1.00
fish 0.00
amphibian 0.00
insect 0.00
mollusc.et.al 0.25Gradient Boosted Decision Trees (xgboost)
xgboostFit <- Zoo_train |> train(type ~ .,
method = "xgbTree",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index),
tuneGrid = expand.grid(
nrounds = 20,
max_depth = 3,
colsample_bytree = .6,
eta = 0.1,
gamma=0,
min_child_weight = 1,
subsample = .5
))
xgboostFiteXtreme Gradient Boosting
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 77, 73, 74, 75, 75, 75, ...
Resampling results:
Accuracy Kappa
0.973 0.964
Tuning parameter 'nrounds' was held constant at a value of 20
Tuning
held constant at a value of 1
Tuning parameter 'subsample' was held
constant at a value of 0.5xgboostFit$finalModel##### xgb.Booster
raw: 112.4 Kb
call:
xgboost::xgb.train(params = list(eta = param$eta, max_depth = param$max_depth,
gamma = param$gamma, colsample_bytree = param$colsample_bytree,
min_child_weight = param$min_child_weight, subsample = param$subsample),
data = x, nrounds = param$nrounds, num_class = length(lev),
objective = "multi:softprob")
params (as set within xgb.train):
eta = "0.1", max_depth = "3", gamma = "0", colsample_bytree = "0.6", min_child_weight = "1", subsample = "0.5", num_class = "7", objective = "multi:softprob", validate_parameters = "TRUE"
xgb.attributes:
niter
callbacks:
cb.print.evaluation(period = print_every_n)
# of features: 16
niter: 20
nfeatures : 16
xNames : hairTRUE feathersTRUE eggsTRUE milkTRUE airborneTRUE aquaticTRUE predatorTRUE toothedTRUE backboneTRUE breathesTRUE venomousTRUE finsTRUE legs tailTRUE domesticTRUE catsizeTRUE
problemType : Classification
tuneValue :
nrounds max_depth eta gamma colsample_bytree min_child_weight subsample
1 20 3 0.1 0 0.6 1 0.5
obsLevels : mammal bird reptile fish amphibian insect mollusc.et.al
param :
list()Artificial Neural Network
nnetFit <- Zoo_train |> train(type ~ .,
method = "nnet",
data = _,
tuneLength = 5,
trControl = trainControl(method = "cv", indexOut = train_index),
trace = FALSE)
nnetFitNeural Network
83 samples
16 predictors
7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 75, 74, 74, 74, 74, 75, ...
Resampling results across tuning parameters:
size decay Accuracy Kappa
1 0e+00 0.776 0.681
1 1e-04 0.789 0.709
1 1e-03 0.911 0.882
1 1e-02 0.832 0.781
1 1e-01 0.722 0.621
3 0e+00 0.963 0.950
3 1e-04 0.976 0.968
3 1e-03 0.986 0.979
3 1e-02 0.986 0.981
3 1e-01 0.976 0.968
5 0e+00 0.965 0.953
5 1e-04 0.986 0.981
5 1e-03 0.986 0.981
5 1e-02 0.986 0.981
5 1e-01 0.986 0.981
7 0e+00 0.976 0.968
7 1e-04 0.986 0.981
7 1e-03 0.986 0.981
7 1e-02 0.986 0.981
7 1e-01 0.986 0.981
9 0e+00 0.986 0.981
9 1e-04 0.986 0.981
9 1e-03 0.986 0.981
9 1e-02 0.986 0.981
9 1e-01 0.986 0.981
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were size = 3 and decay = 0.01.nnetFit$finalModela 16-3-7 network with 79 weights
inputs: hairTRUE feathersTRUE eggsTRUE milkTRUE airborneTRUE aquaticTRUE predatorTRUE toothedTRUE backboneTRUE breathesTRUE venomousTRUE finsTRUE legs tailTRUE domesticTRUE catsizeTRUE
output(s): .outcome
options were - softmax modelling decay=0.01Comparing Models
Collect the performance metrics from the models trained on the same data.
resamps <- resamples(list(
ctree = ctreeFit,
C45 = C45Fit,
SVM = svmFit,
KNN = knnFit,
rules = rulesFit,
randomForest = randomForestFit,
xgboost = xgboostFit,
NeuralNet = nnetFit
))
resamps
Call:
resamples.default(x = list(ctree = ctreeFit, C45 = C45Fit, SVM = svmFit, KNN
= knnFit, rules = rulesFit, randomForest = randomForestFit, xgboost
= xgboostFit, NeuralNet = nnetFit))
Models: ctree, C45, SVM, KNN, rules, randomForest, xgboost, NeuralNet
Number of resamples: 10
Performance metrics: Accuracy, Kappa
Time estimates for: everything, final model fit Calculate summary statistics
summary(resamps)
Call:
summary.resamples(object = resamps)
Models: ctree, C45, SVM, KNN, rules, randomForest, xgboost, NeuralNet
Number of resamples: 10
Accuracy
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
ctree 0.700 0.778 0.817 0.827 0.871 1 0
C45 1.000 1.000 1.000 1.000 1.000 1 0
SVM 1.000 1.000 1.000 1.000 1.000 1 0
KNN 1.000 1.000 1.000 1.000 1.000 1 0
rules 0.900 1.000 1.000 0.990 1.000 1 0
randomForest 1.000 1.000 1.000 1.000 1.000 1 0
xgboost 0.857 1.000 1.000 0.973 1.000 1 0
NeuralNet 0.857 1.000 1.000 0.986 1.000 1 0
Kappa
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
ctree 0.634 0.715 0.748 0.772 0.815 1 0
C45 1.000 1.000 1.000 1.000 1.000 1 0
SVM 1.000 1.000 1.000 1.000 1.000 1 0
KNN 1.000 1.000 1.000 1.000 1.000 1 0
rules 0.868 1.000 1.000 0.987 1.000 1 0
randomForest 1.000 1.000 1.000 1.000 1.000 1 0
xgboost 0.806 1.000 1.000 0.964 1.000 1 0
NeuralNet 0.806 1.000 1.000 0.981 1.000 1 0library(lattice)
bwplot(resamps, layout = c(3, 1))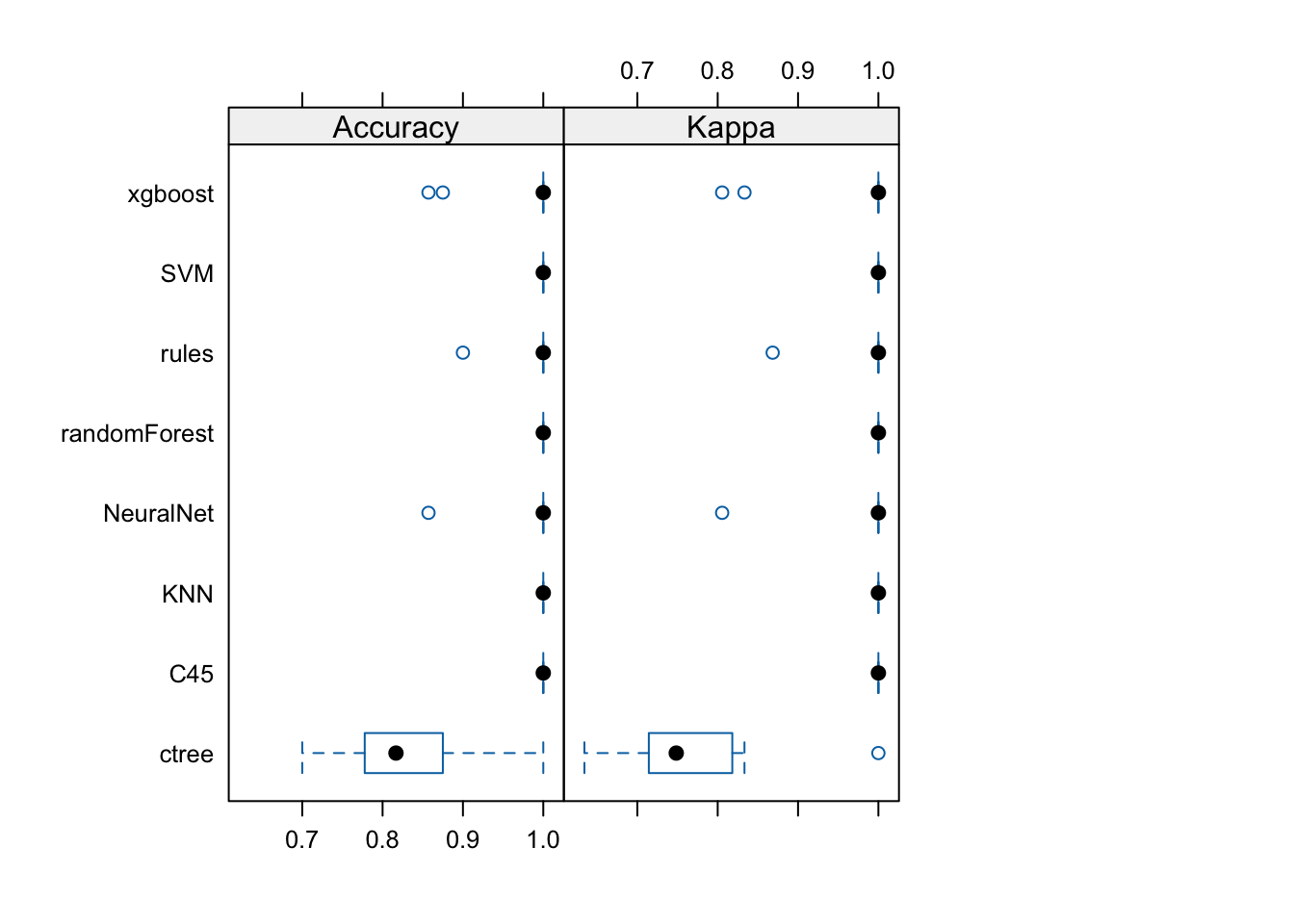
Perform inference about differences between models. For each metric, all pair-wise differences are computed and tested to assess if the difference is equal to zero. By default Bonferroni correction for multiple comparison is used. Differences are shown in the upper triangle and p-values are in the lower triangle.
difs <- diff(resamps)
difs
Call:
diff.resamples(x = resamps)
Models: ctree, C45, SVM, KNN, rules, randomForest, xgboost, NeuralNet
Metrics: Accuracy, Kappa
Number of differences: 28
p-value adjustment: bonferroni summary(difs)
Call:
summary.diff.resamples(object = difs)
p-value adjustment: bonferroni
Upper diagonal: estimates of the difference
Lower diagonal: p-value for H0: difference = 0
Accuracy
ctree C45 SVM KNN rules randomForest xgboost
ctree -0.17262 -0.17262 -0.17262 -0.16262 -0.17262 -0.14583
C45 0.00193 0.00000 0.00000 0.01000 0.00000 0.02679
SVM 0.00193 NA 0.00000 0.01000 0.00000 0.02679
KNN 0.00193 NA NA 0.01000 0.00000 0.02679
rules 0.00376 1.00000 1.00000 1.00000 -0.01000 0.01679
randomForest 0.00193 NA NA NA 1.00000 0.02679
xgboost 0.05129 1.00000 1.00000 1.00000 1.00000 1.00000
NeuralNet 0.01405 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
NeuralNet
ctree -0.15833
C45 0.01429
SVM 0.01429
KNN 0.01429
rules 0.00429
randomForest 0.01429
xgboost -0.01250
NeuralNet
Kappa
ctree C45 SVM KNN rules randomForest xgboost
ctree -0.22840 -0.22840 -0.22840 -0.21524 -0.22840 -0.19229
C45 0.00116 0.00000 0.00000 0.01316 0.00000 0.03611
SVM 0.00116 NA 0.00000 0.01316 0.00000 0.03611
KNN 0.00116 NA NA 0.01316 0.00000 0.03611
rules 0.00238 1.00000 1.00000 1.00000 -0.01316 0.02295
randomForest 0.00116 NA NA NA 1.00000 0.03611
xgboost 0.04216 1.00000 1.00000 1.00000 1.00000 1.00000
NeuralNet 0.01055 1.00000 1.00000 1.00000 1.00000 1.00000 1.00000
NeuralNet
ctree -0.20895
C45 0.01944
SVM 0.01944
KNN 0.01944
rules 0.00629
randomForest 0.01944
xgboost -0.01667
NeuralNet All perform similarly well except ctree (differences in the first row are negative and the p-values in the first column are <.05 indicating that the null-hypothesis of a difference of 0 can be rejected).
Applying the Chosen Model to the Test Data
Most models do similarly well on the data. We choose here the random forest model.
pr <- predict(randomForestFit, Zoo_test)
pr [1] mammal mammal mammal fish fish
[6] bird bird mammal mammal mammal
[11] mammal mollusc.et.al reptile mammal bird
[16] mollusc.et.al bird insect
Levels: mammal bird reptile fish amphibian insect mollusc.et.alCalculate the confusion matrix for the held-out test data.
confusionMatrix(pr, reference = Zoo_test$type)Confusion Matrix and Statistics
Reference
Prediction mammal bird reptile fish amphibian insect mollusc.et.al
mammal 8 0 0 0 0 0 0
bird 0 4 0 0 0 0 0
reptile 0 0 1 0 0 0 0
fish 0 0 0 2 0 0 0
amphibian 0 0 0 0 0 0 0
insect 0 0 0 0 0 1 0
mollusc.et.al 0 0 0 0 0 0 2
Overall Statistics
Accuracy : 1
95% CI : (0.815, 1)
No Information Rate : 0.444
P-Value [Acc > NIR] : 4.58e-07
Kappa : 1
Mcnemar's Test P-Value : NA
Statistics by Class:
Class: mammal Class: bird Class: reptile Class: fish
Sensitivity 1.000 1.000 1.0000 1.000
Specificity 1.000 1.000 1.0000 1.000
Pos Pred Value 1.000 1.000 1.0000 1.000
Neg Pred Value 1.000 1.000 1.0000 1.000
Prevalence 0.444 0.222 0.0556 0.111
Detection Rate 0.444 0.222 0.0556 0.111
Detection Prevalence 0.444 0.222 0.0556 0.111
Balanced Accuracy 1.000 1.000 1.0000 1.000
Class: amphibian Class: insect Class: mollusc.et.al
Sensitivity NA 1.0000 1.000
Specificity 1 1.0000 1.000
Pos Pred Value NA 1.0000 1.000
Neg Pred Value NA 1.0000 1.000
Prevalence 0 0.0556 0.111
Detection Rate 0 0.0556 0.111
Detection Prevalence 0 0.0556 0.111
Balanced Accuracy NA 1.0000 1.000Comparing Decision Boundaries of Popular Classification Techniques
Classifiers create decision boundaries to discriminate between classes. Different classifiers are able to create different shapes of decision boundaries (e.g., some are strictly linear) and thus some classifiers may perform better for certain datasets. This page visualizes the decision boundaries found by several popular classification methods.
The following plot adds the decision boundary (black lines) and classification confidence (color intensity) by evaluating the classifier at evenly spaced grid points. Note that low resolution (to make evaluation faster) will make the decision boundary look like it has small steps even if it is a (straight) line.
library(scales)
library(tidyverse)
library(ggplot2)
library(caret)
decisionplot <- function(model, data, class_var,
predict_type = c("class", "prob"), resolution = 3 * 72) {
# resolution is set to 72 dpi if the image is rendered 3 inches wide.
y <- data |> pull(class_var)
x <- data |> dplyr::select(-all_of(class_var))
# resubstitution accuracy
prediction <- predict(model, x, type = predict_type[1])
# LDA returns a list
if(is.list(prediction)) prediction <- prediction$class
prediction <- factor(prediction, levels = levels(y))
cm <- confusionMatrix(data = prediction,
reference = y)
acc <- cm$overall["Accuracy"]
# evaluate model on a grid
r <- sapply(x[, 1:2], range, na.rm = TRUE)
xs <- seq(r[1,1], r[2,1], length.out = resolution)
ys <- seq(r[1,2], r[2,2], length.out = resolution)
g <- cbind(rep(xs, each = resolution), rep(ys, time = resolution))
colnames(g) <- colnames(r)
g <- as_tibble(g)
### guess how to get class labels from predict
### (unfortunately not very consistent between models)
cl <- predict(model, g, type = predict_type[1])
# LDA returns a list
prob <- NULL
if(is.list(cl)) {
prob <- cl$posterior
cl <- cl$class
} else
if(!is.na(predict_type[2]))
try(prob <- predict(model, g, type = predict_type[2]))
# we visualize the difference in probability/score between the
# winning class and the second best class.
# don't use probability if predict for the classifier does not support it.
max_prob <- 1
if(!is.null(prob))
try({
max_prob <- t(apply(prob, MARGIN = 1, sort, decreasing = TRUE))
max_prob <- max_prob[,1] - max_prob[,2]
}, silent = TRUE)
cl <- factor(cl, levels = levels(y))
g <- g |> add_column(prediction = cl, probability = max_prob)
ggplot(g, mapping = aes(
x = .data[[colnames(g)[1]]], y = .data[[colnames(g)[2]]])) +
geom_raster(mapping = aes(fill = prediction, alpha = probability)) +
geom_contour(mapping = aes(z = as.numeric(prediction)),
bins = length(levels(cl)), linewidth = .5, color = "black") +
geom_point(data = data, mapping = aes(
x = .data[[colnames(data)[1]]],
y = .data[[colnames(data)[2]]],
shape = .data[[class_var]]), alpha = .7) +
scale_alpha_continuous(range = c(0,1), limits = c(0,1), guide = "none") +
labs(subtitle = paste("Training accuracy:", round(acc, 2))) +
theme_minimal(base_size = 14)
}Penguins Dataset
For easier visualization, we use two dimensions of the penguins dataset. Contour lines visualize the density like mountains on a map.
set.seed(1000)
data("penguins")
penguins <- as_tibble(penguins) |>
drop_na()
### Three classes
### (note: MASS also has a select function which hides dplyr's select)
x <- penguins |> dplyr::select(bill_length_mm, bill_depth_mm, species)
x# A tibble: 333 × 3
bill_length_mm bill_depth_mm species
<dbl> <dbl> <fct>
1 39.1 18.7 Adelie
2 39.5 17.4 Adelie
3 40.3 18 Adelie
4 36.7 19.3 Adelie
5 39.3 20.6 Adelie
6 38.9 17.8 Adelie
7 39.2 19.6 Adelie
8 41.1 17.6 Adelie
9 38.6 21.2 Adelie
10 34.6 21.1 Adelie
# ℹ 323 more rowsggplot(x, aes(x = bill_length_mm, y = bill_depth_mm, fill = species)) +
stat_density_2d(geom = "polygon", aes(alpha = after_stat(level))) +
geom_point() +
theme_minimal(base_size = 14) +
labs(x = "Bill length (mm)",
y = "Bill depth (mm)",
fill = "Species",
alpha = "Density")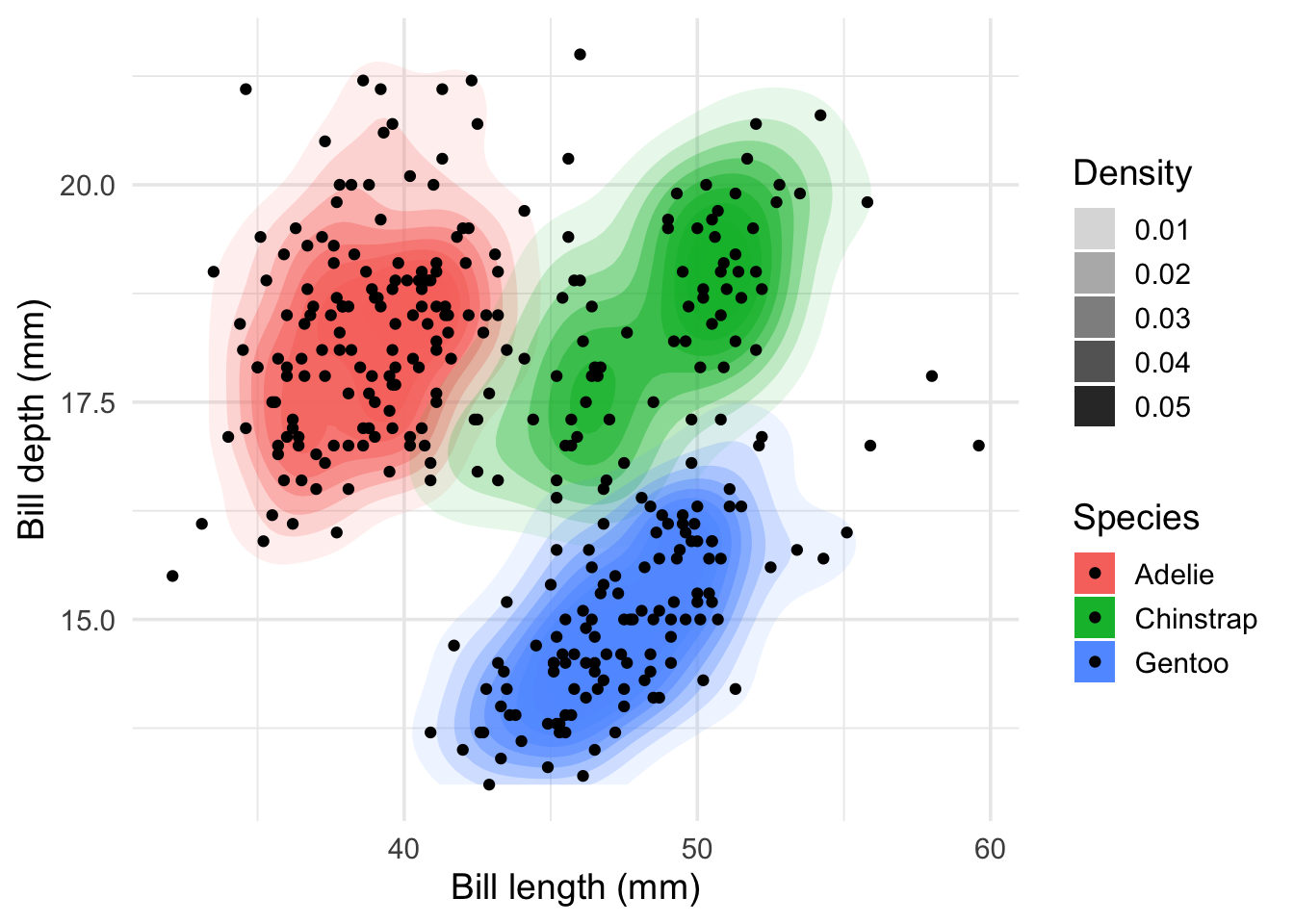
Note: There is some overplotting and you could use geom_jitter() instead of geom_point().
K-Nearest Neighbors Classifier
model <- x |> caret::knn3(species ~ ., data = _, k = 1)
decisionplot(model, x, class_var = "species") +
labs(title = "kNN (1 neighbor)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")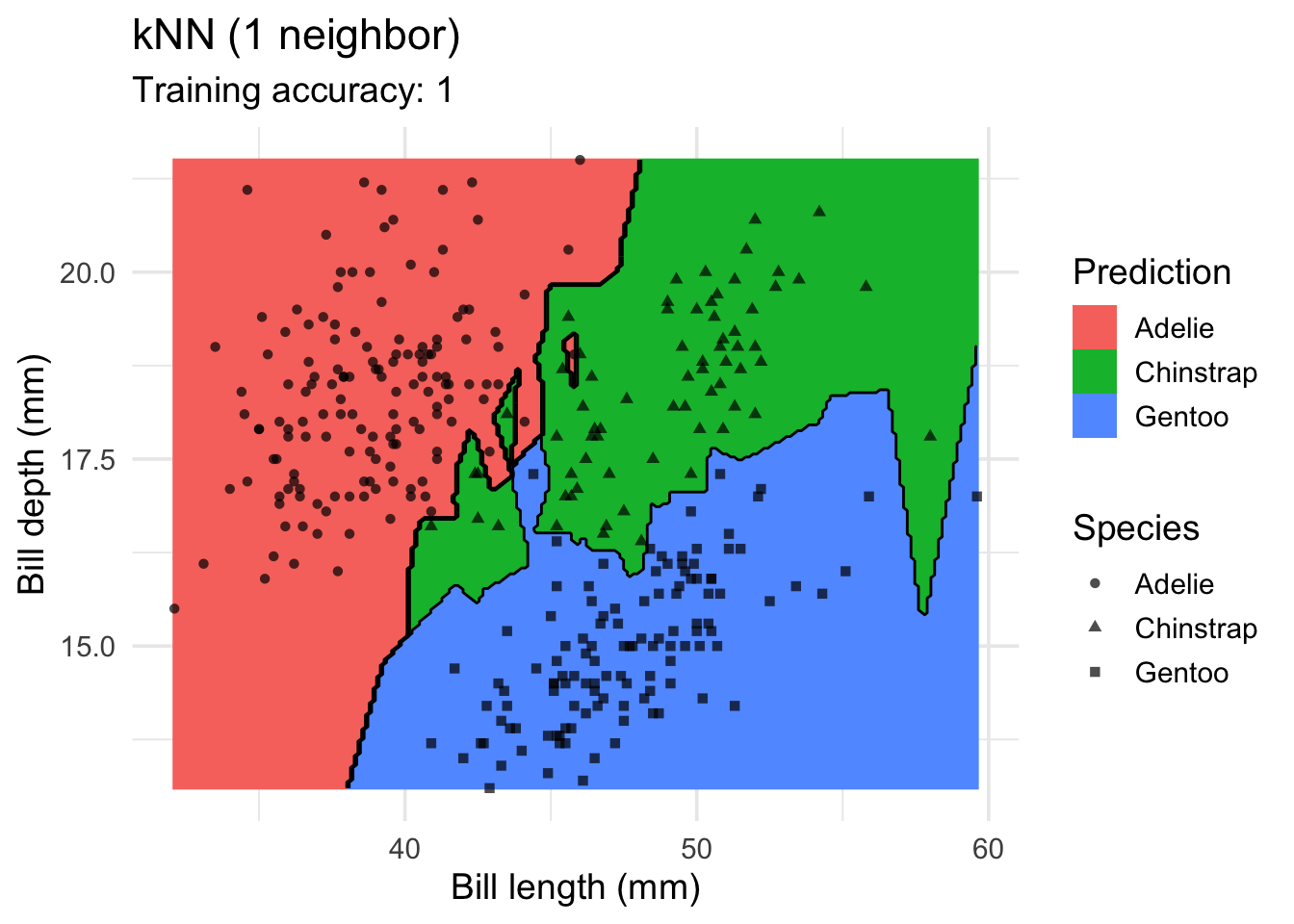
model <- x |> caret::knn3(species ~ ., data = _, k = 3)
decisionplot(model, x, class_var = "species") +
labs(title = "kNN (3 neighbor)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")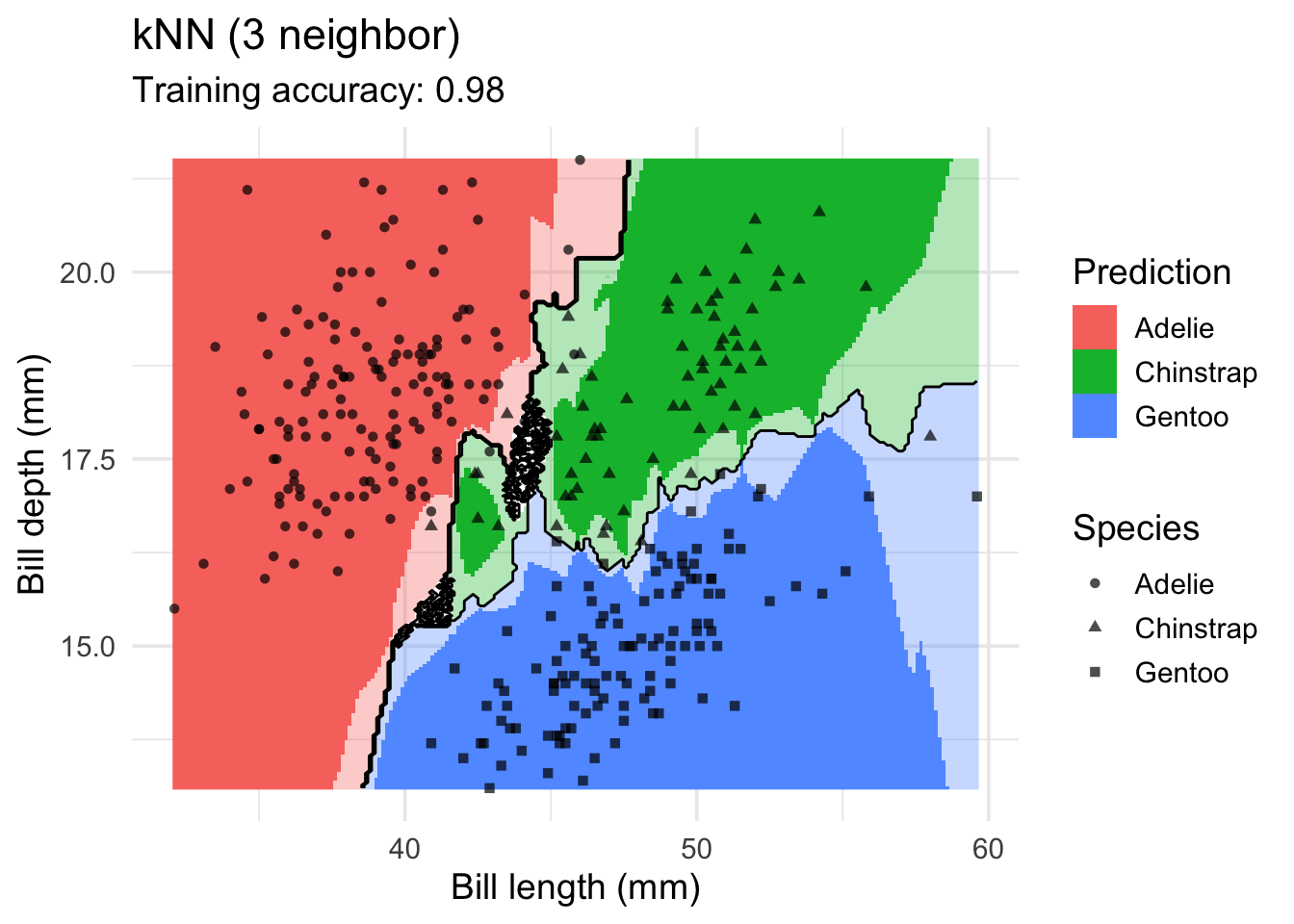
model <- x |> caret::knn3(species ~ ., data = _, k = 9)
decisionplot(model, x, class_var = "species") +
labs(title = "kNN (9 neighbor)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")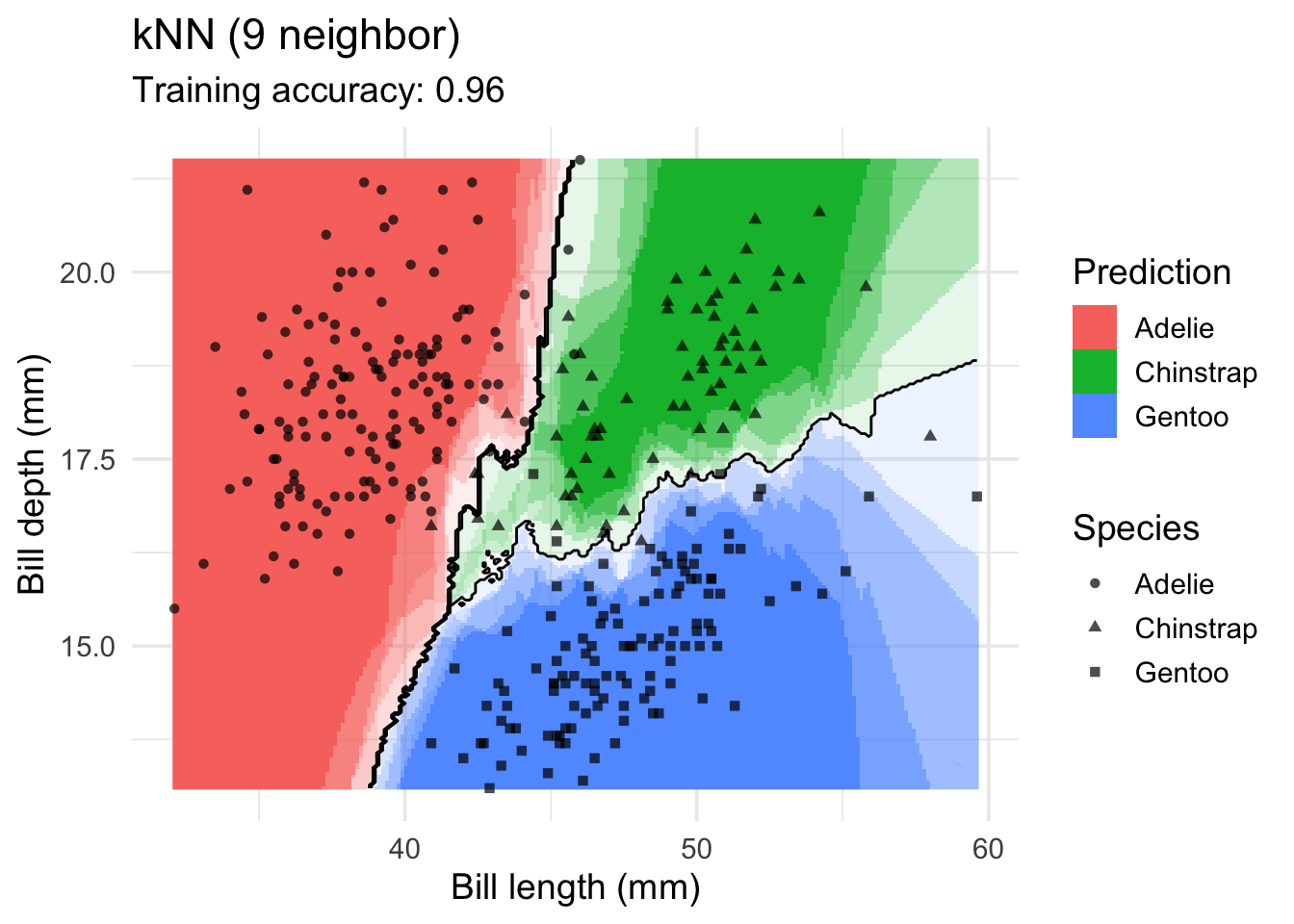
Increasing \(k\) smooths the decision boundary. At \(k=1\), we see white areas around points where penguins of two classes are in the same spot. Here, the algorithm randomly chooses a class during prediction resulting in the meandering decision boundary. The predictions in that area are not stable and every time we ask for a class, we may get a different class.
Naive Bayes Classifier
model <- x |> e1071::naiveBayes(species ~ ., data = _)
decisionplot(model, x, class_var = "species",
predict_type = c("class", "raw")) +
labs(title = "Naive Bayes",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction") 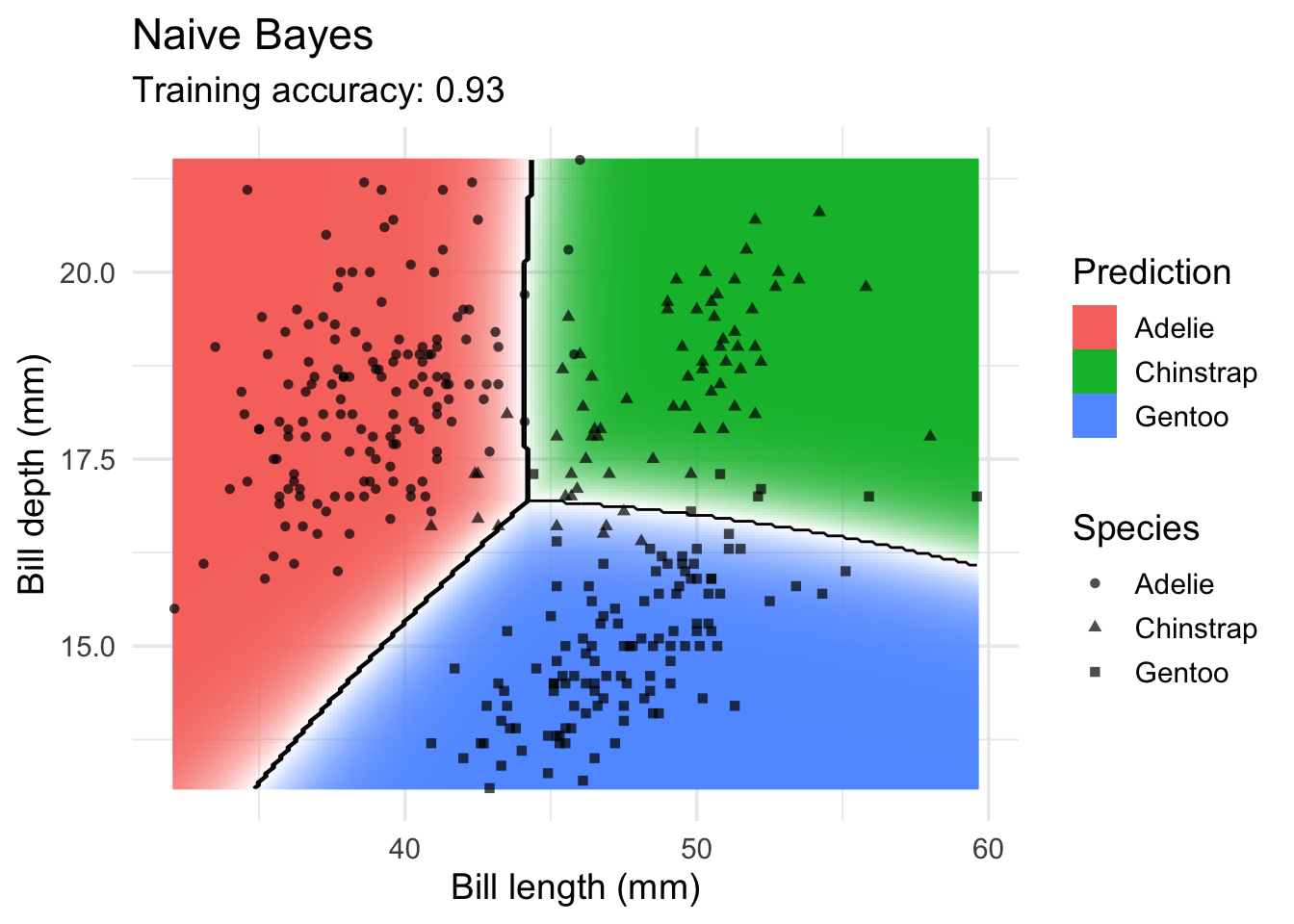
Linear Discriminant Analysis
model <- x |> MASS::lda(species ~ ., data = _)
decisionplot(model, x, class_var = "species") +
labs(title = "LDA",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")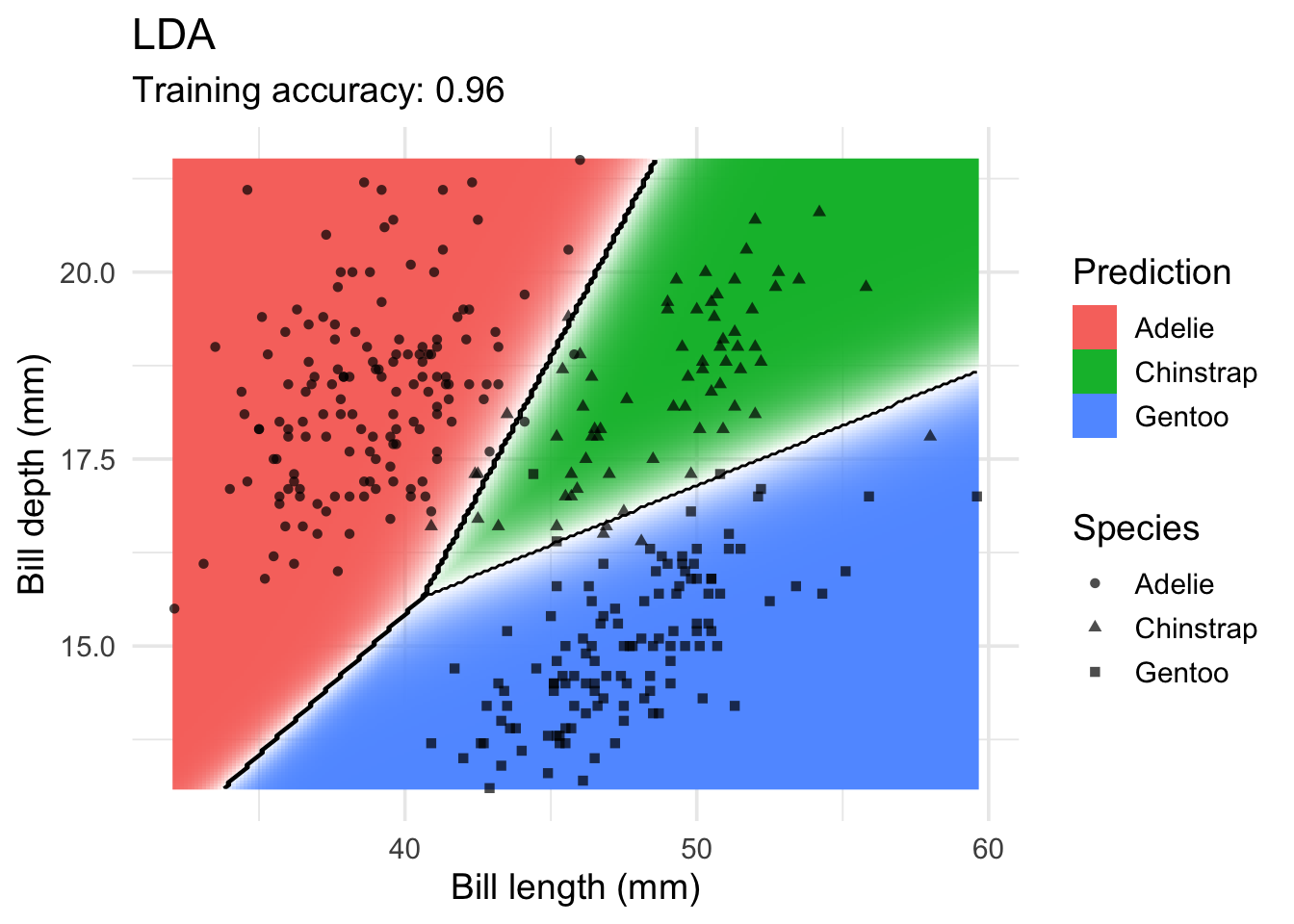
Multinomial Logistic Regression (implemented in nnet)
Multinomial logistic regression is an extension of logistic regression to problems with more than two classes.
model <- x |> nnet::multinom(species ~., data = _)# weights: 12 (6 variable)
initial value 365.837892
iter 10 value 26.650783
iter 20 value 23.943597
iter 30 value 23.916873
iter 40 value 23.901339
iter 50 value 23.895442
iter 60 value 23.894251
final value 23.892065
convergeddecisionplot(model, x, class_var = "species") +
labs(title = "Multinomial Logistic Regression",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")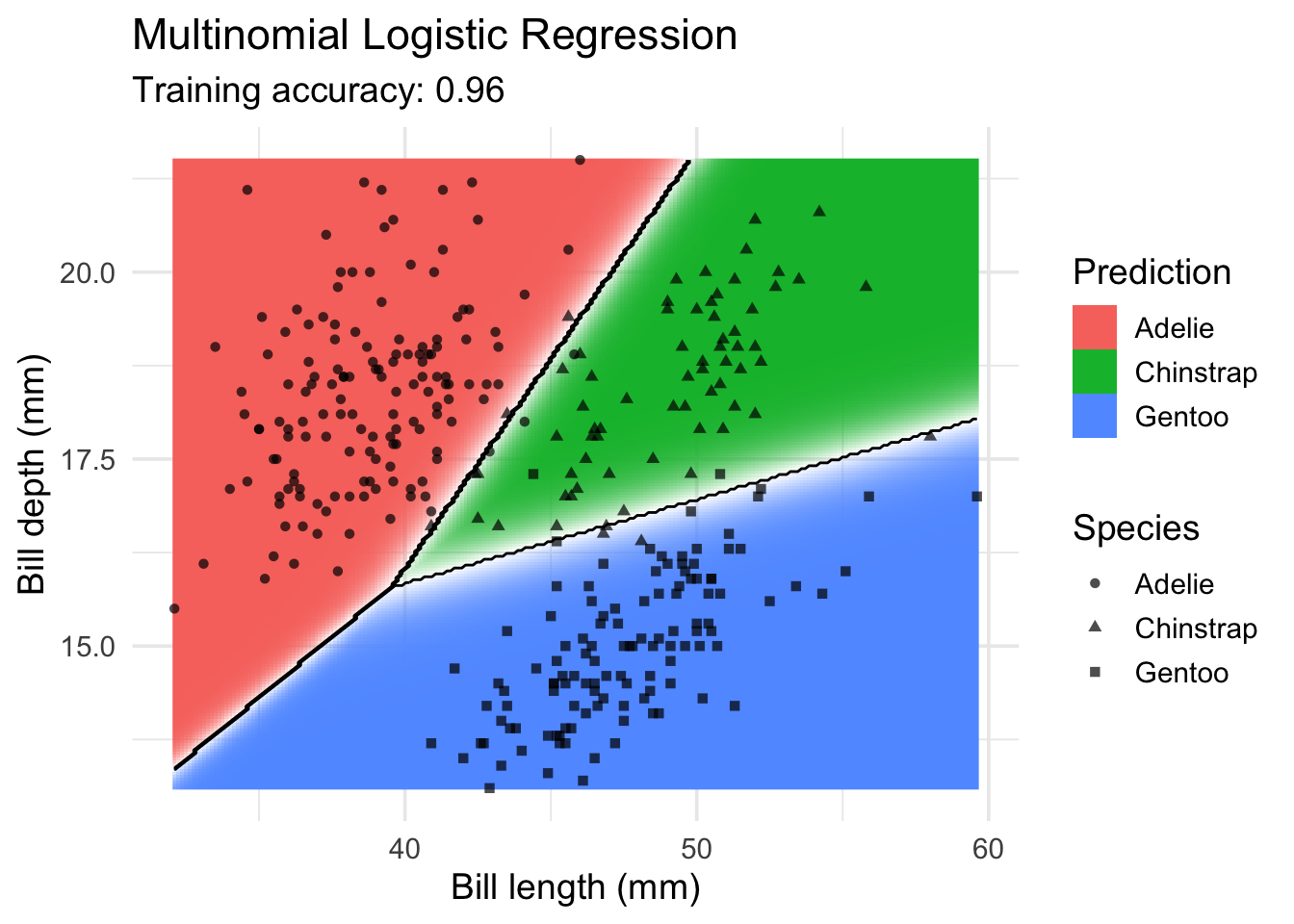
Decision Trees
model <- x |> rpart::rpart(species ~ ., data = _)
decisionplot(model, x, class_var = "species") +
labs(title = "CART",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")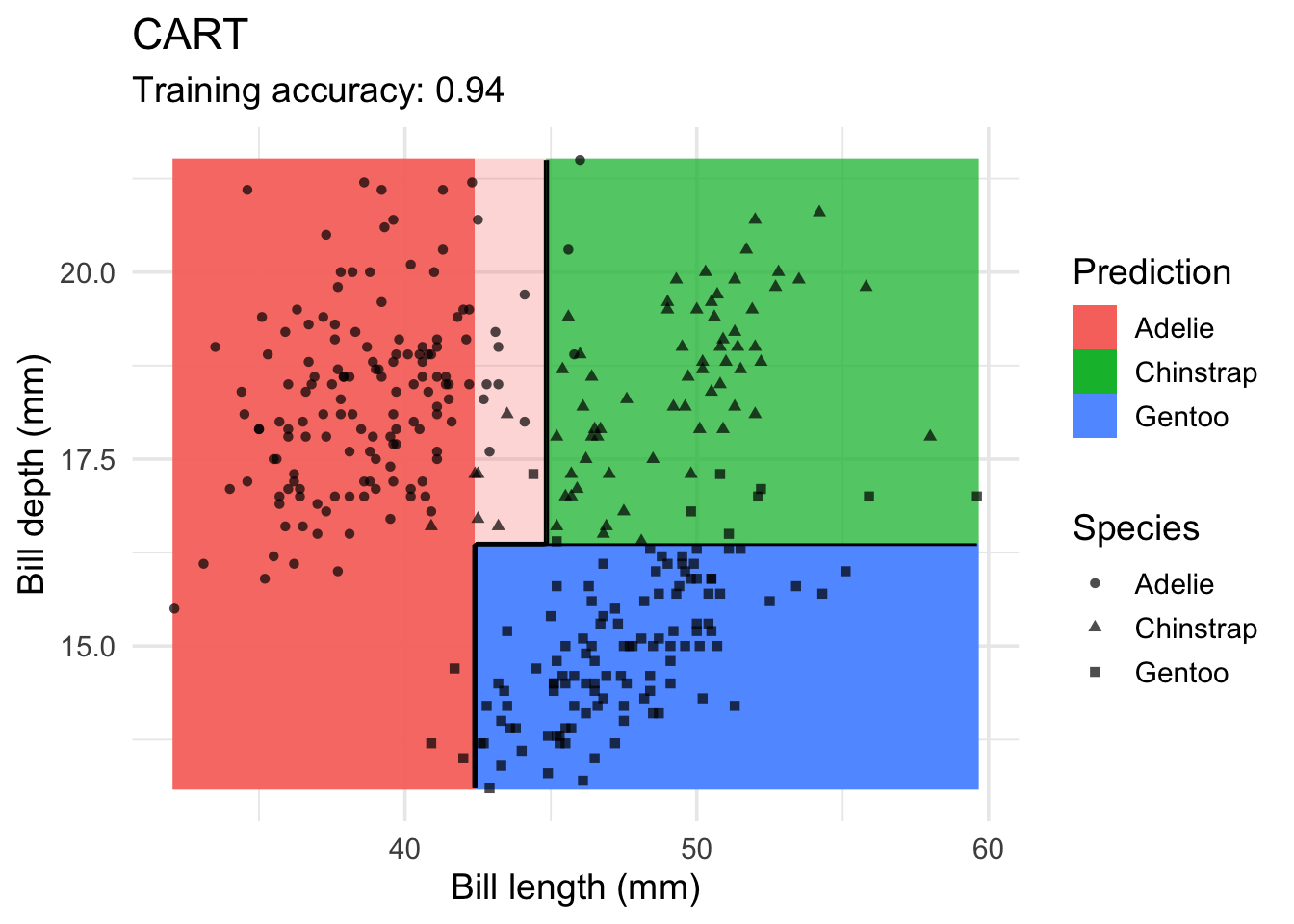
model <- x |> rpart::rpart(species ~ ., data = _,
control = rpart.control(cp = 0.001, minsplit = 1))
decisionplot(model, x, class_var = "species") +
labs(title = "CART (overfitting)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")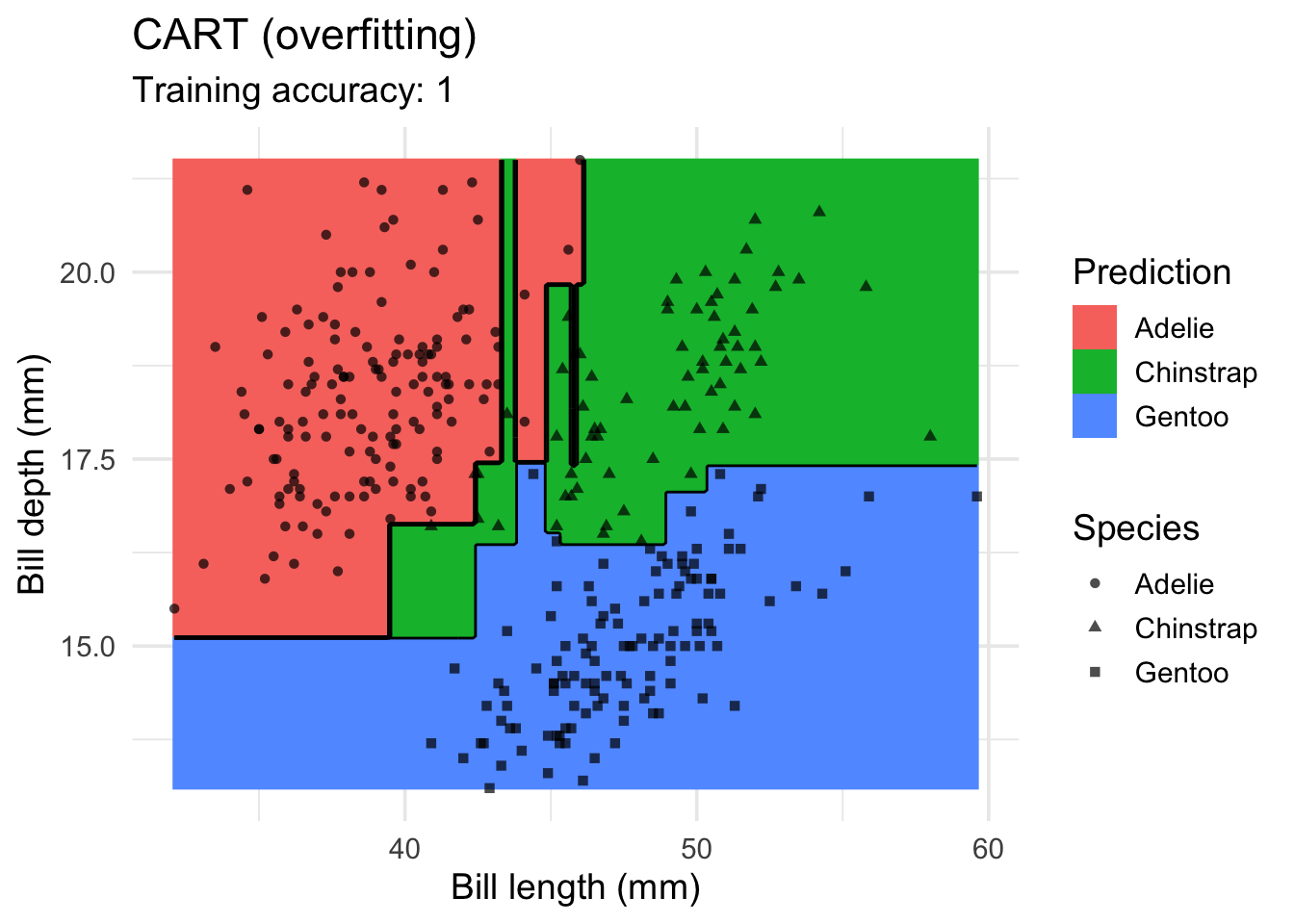
model <- x |> C50::C5.0(species ~ ., data = _)
decisionplot(model, x, class_var = "species") +
labs(title = "C5.0",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")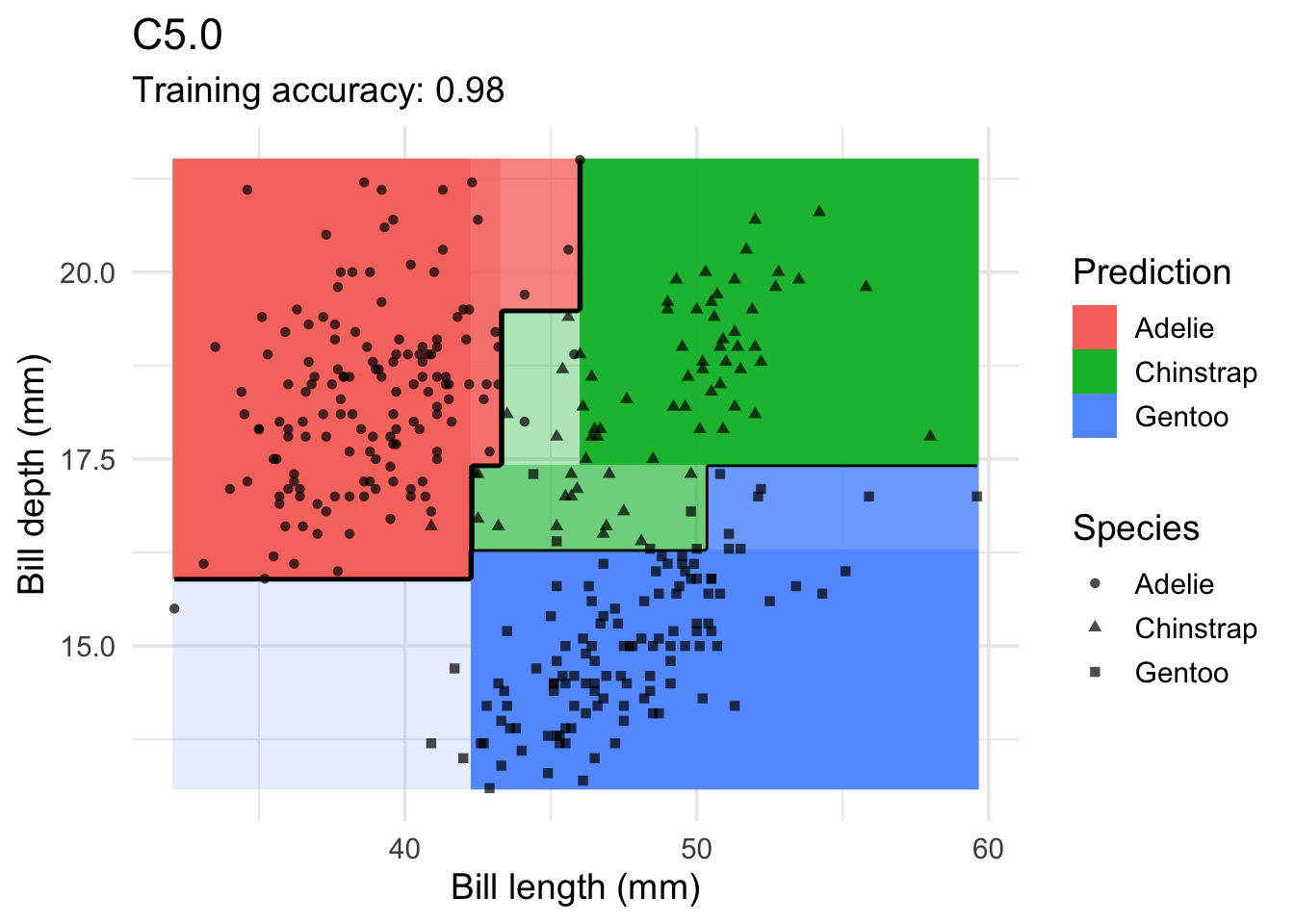
model <- x |> randomForest::randomForest(species ~ ., data = _)
decisionplot(model, x, class_var = "species") +
labs(title = "Random Forest",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")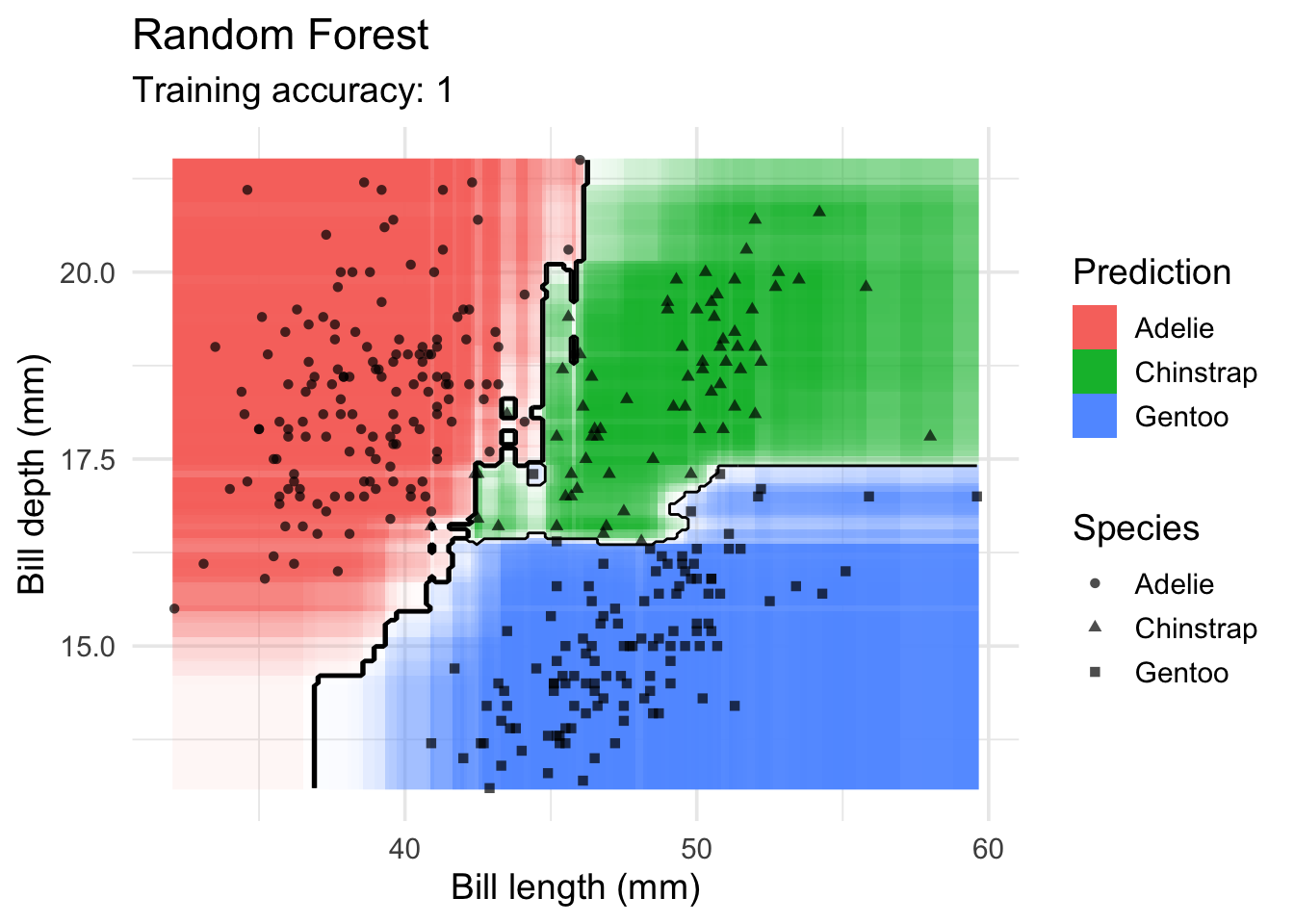
SVM
model <- x |> e1071::svm(species ~ ., data = _, kernel = "linear")
decisionplot(model, x, class_var = "species") +
labs(title = "SVM (linear kernel)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")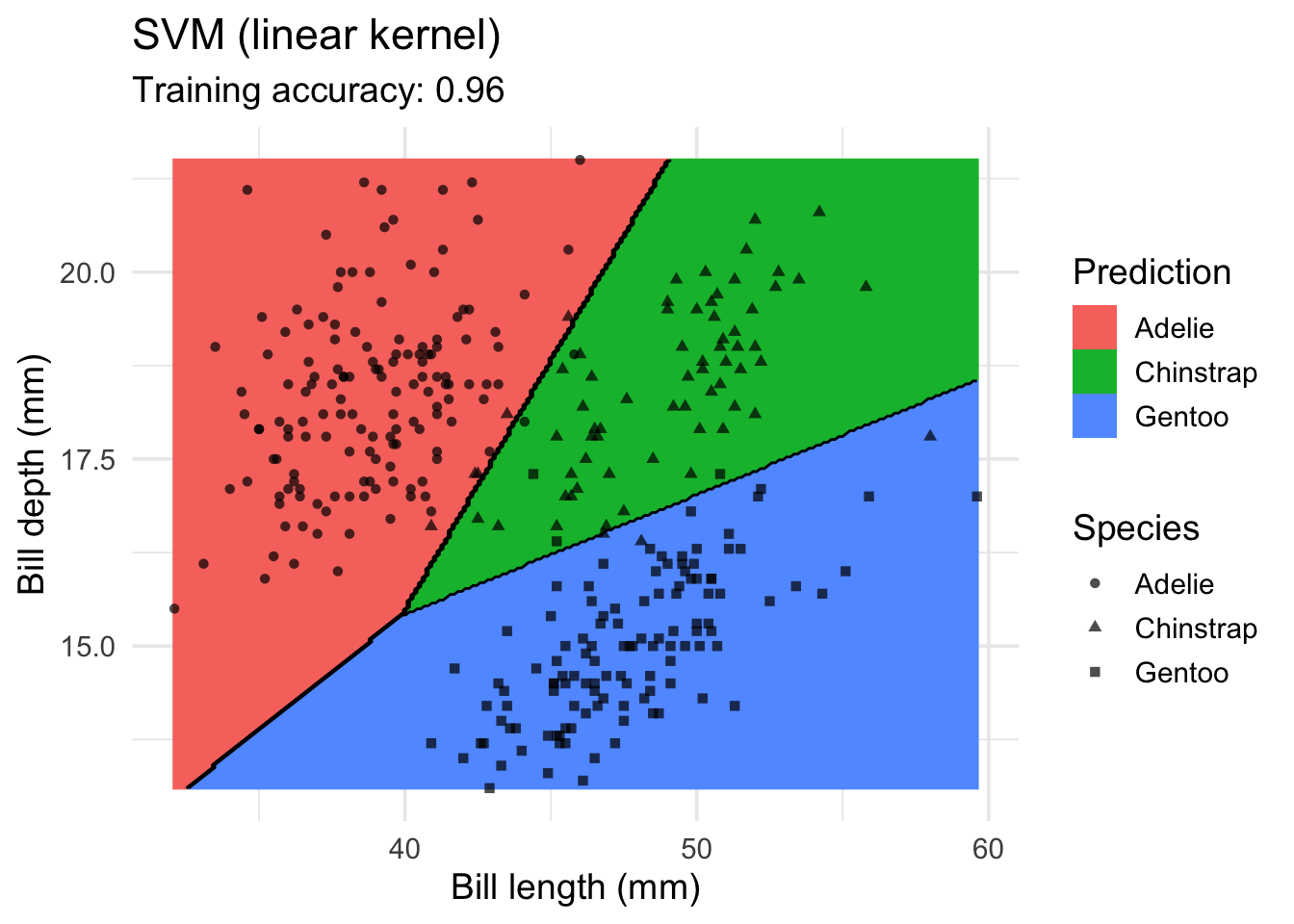
model <- x |> e1071::svm(species ~ ., data = _, kernel = "radial")
decisionplot(model, x, class_var = "species") +
labs(title = "SVM (radial kernel)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")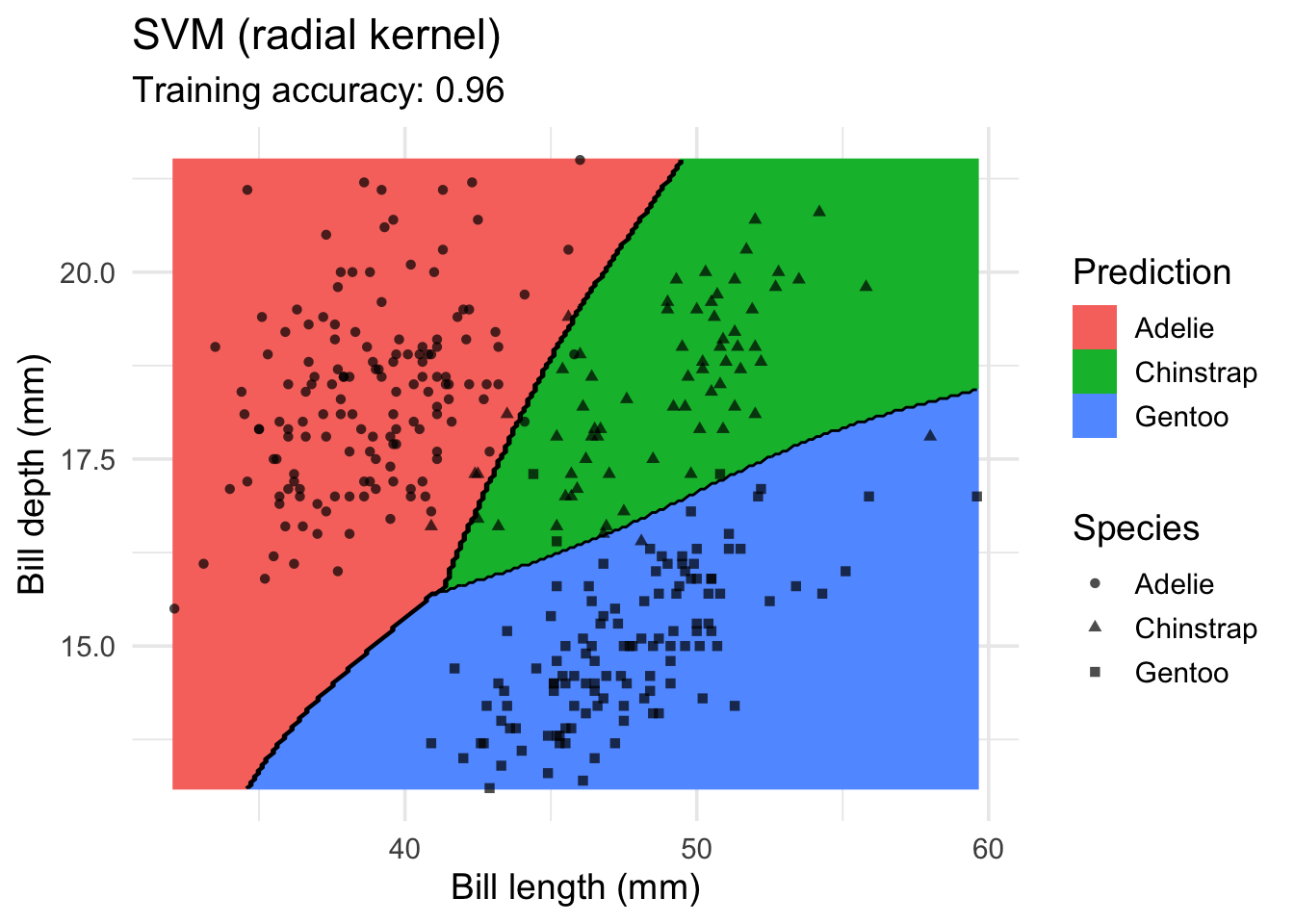
model <- x |> e1071::svm(species ~ ., data = _, kernel = "polynomial")
decisionplot(model, x, class_var = "species") +
labs(title = "SVM (polynomial kernel)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")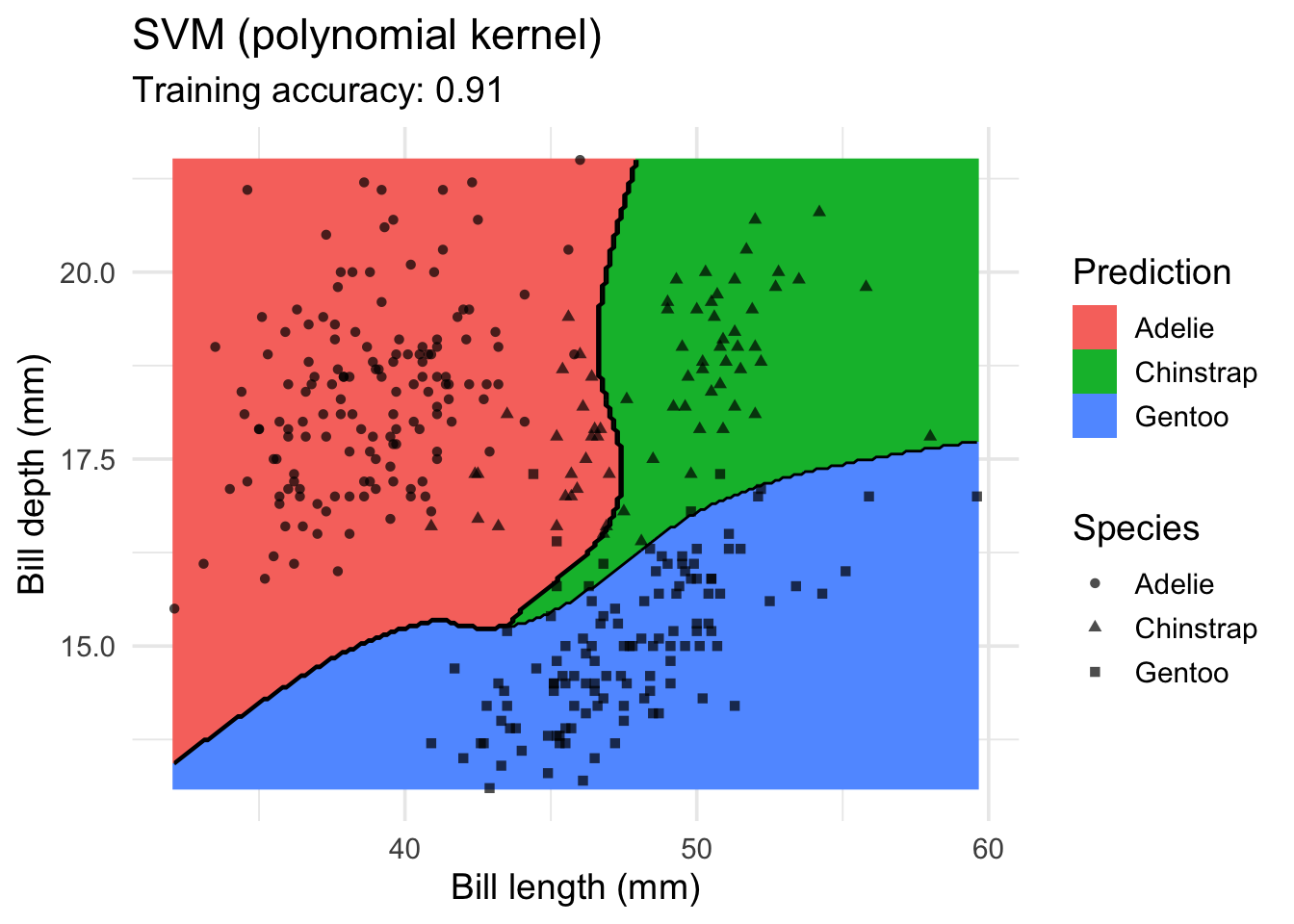
model <- x |> e1071::svm(species ~ ., data = _, kernel = "sigmoid")
decisionplot(model, x, class_var = "species") +
labs(title = "SVM (sigmoid kernel)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")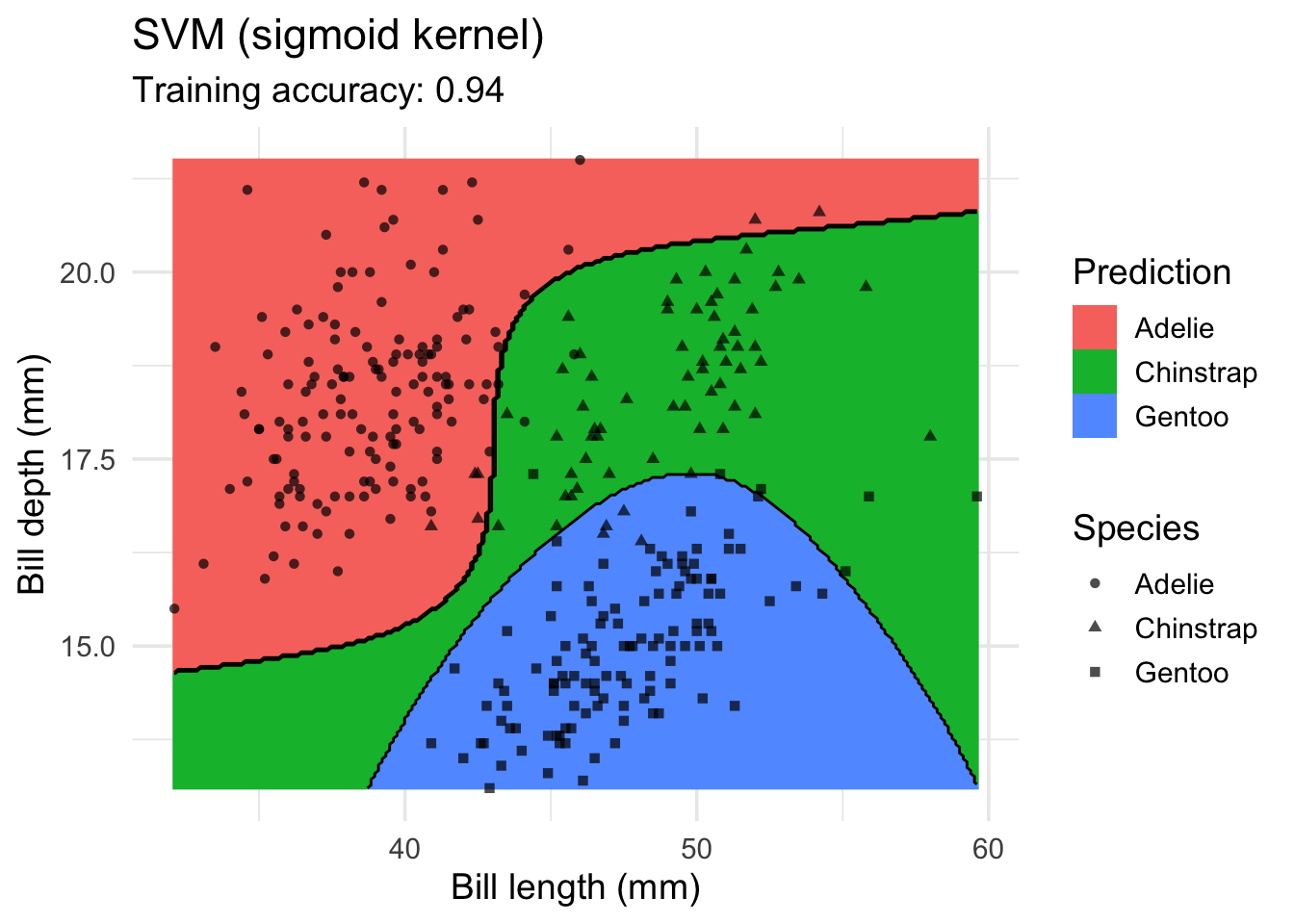
Single Layer Feed-forward Neural Networks
model <-x |> nnet::nnet(species ~ ., data = _, size = 1, trace = FALSE)
decisionplot(model, x, class_var = "species",
predict_type = c("class", "raw")) +
labs(title = "NN (1 neuron)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")Warning: Computation failed in `stat_contour()`
Caused by error in `if (zero_range(range)) ...`:
! missing value where TRUE/FALSE needed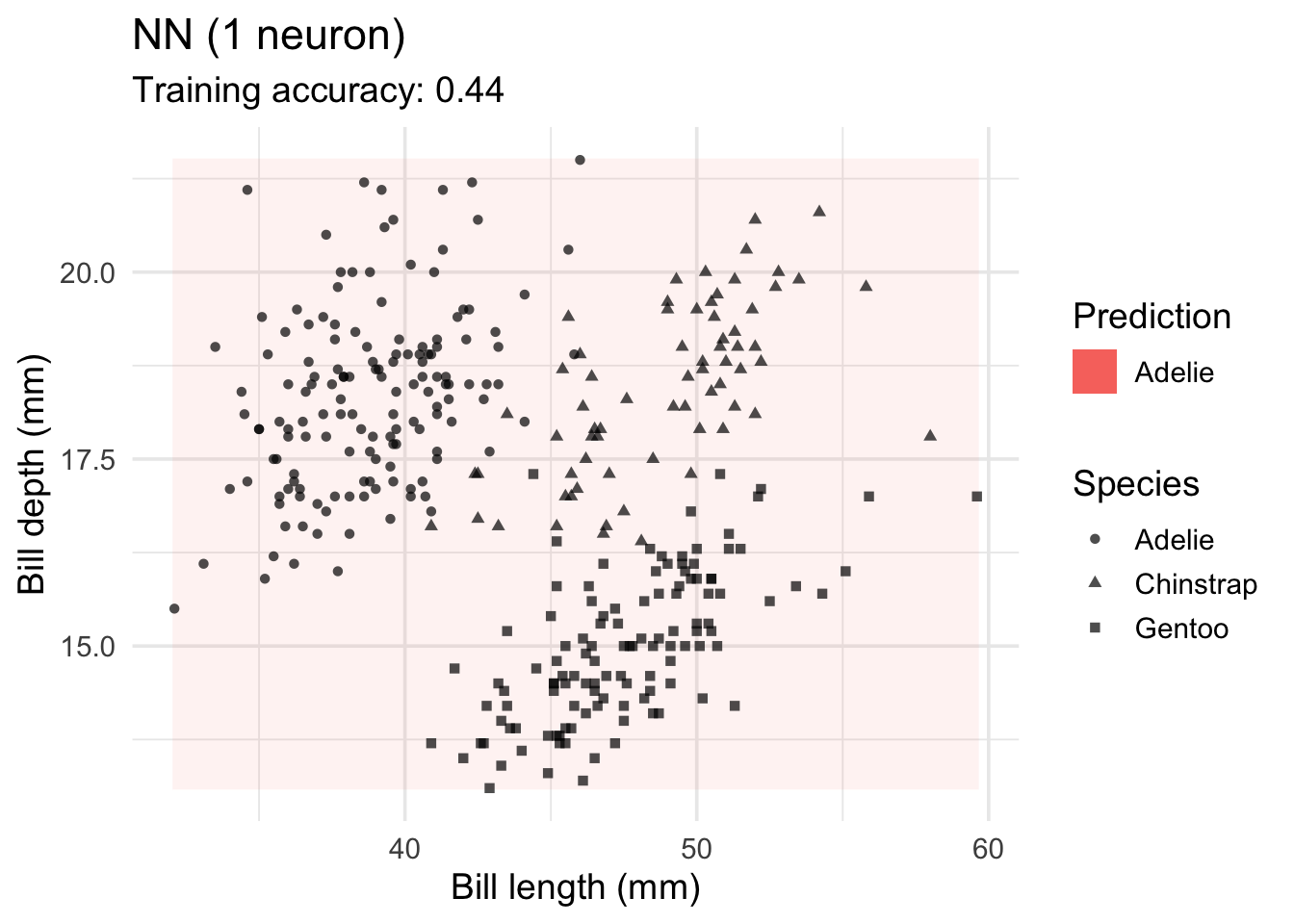
model <-x |> nnet::nnet(species ~ ., data = _, size = 2, trace = FALSE)
decisionplot(model, x, class_var = "species",
predict_type = c("class", "raw")) +
labs(title = "NN (2 neurons)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")Warning: Computation failed in `stat_contour()`
Caused by error in `if (zero_range(range)) ...`:
! missing value where TRUE/FALSE needed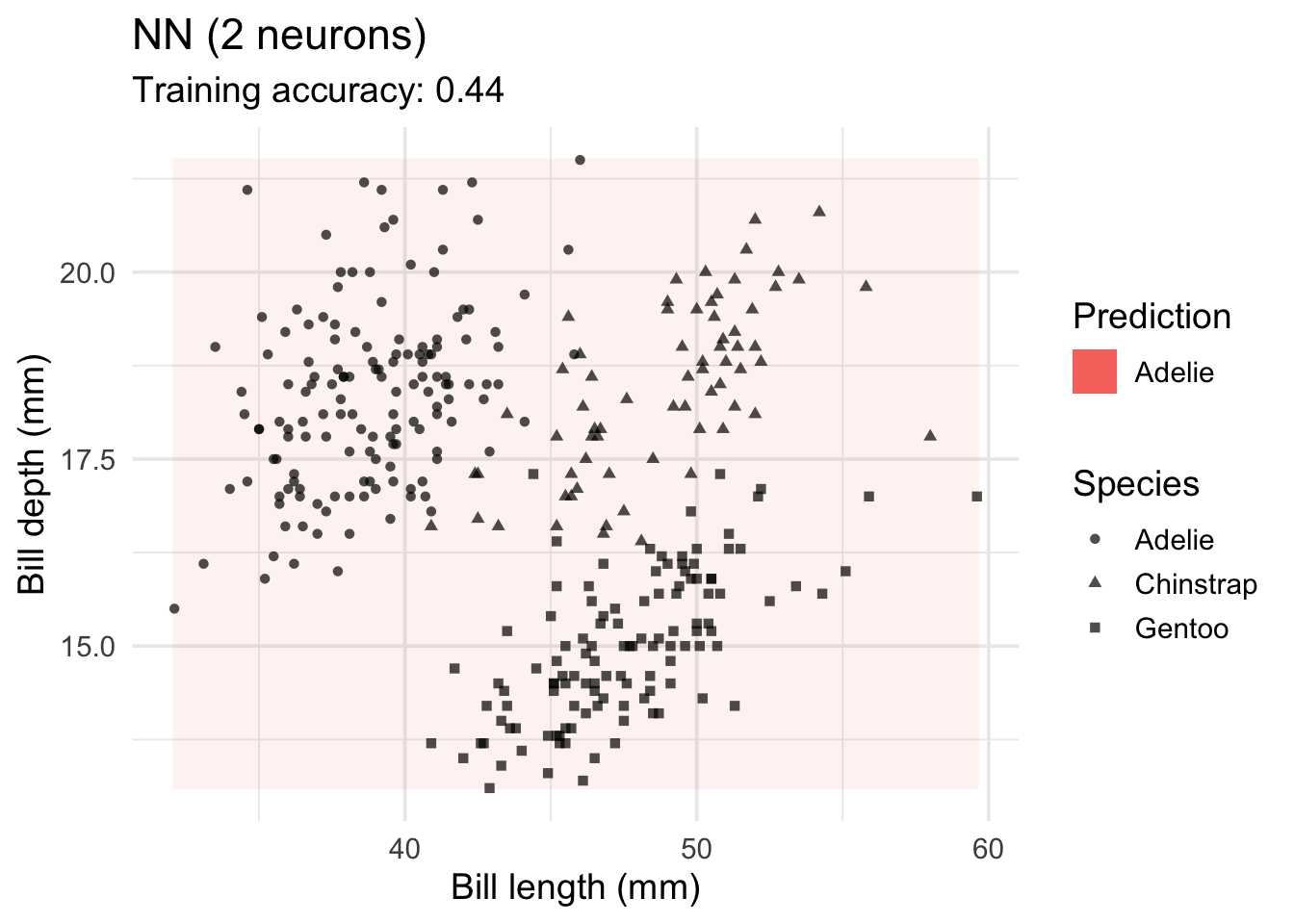
model <-x |> nnet::nnet(species ~ ., data = _, size = 4, trace = FALSE)
decisionplot(model, x, class_var = "species",
predict_type = c("class", "raw")) +
labs(title = "NN (4 neurons)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")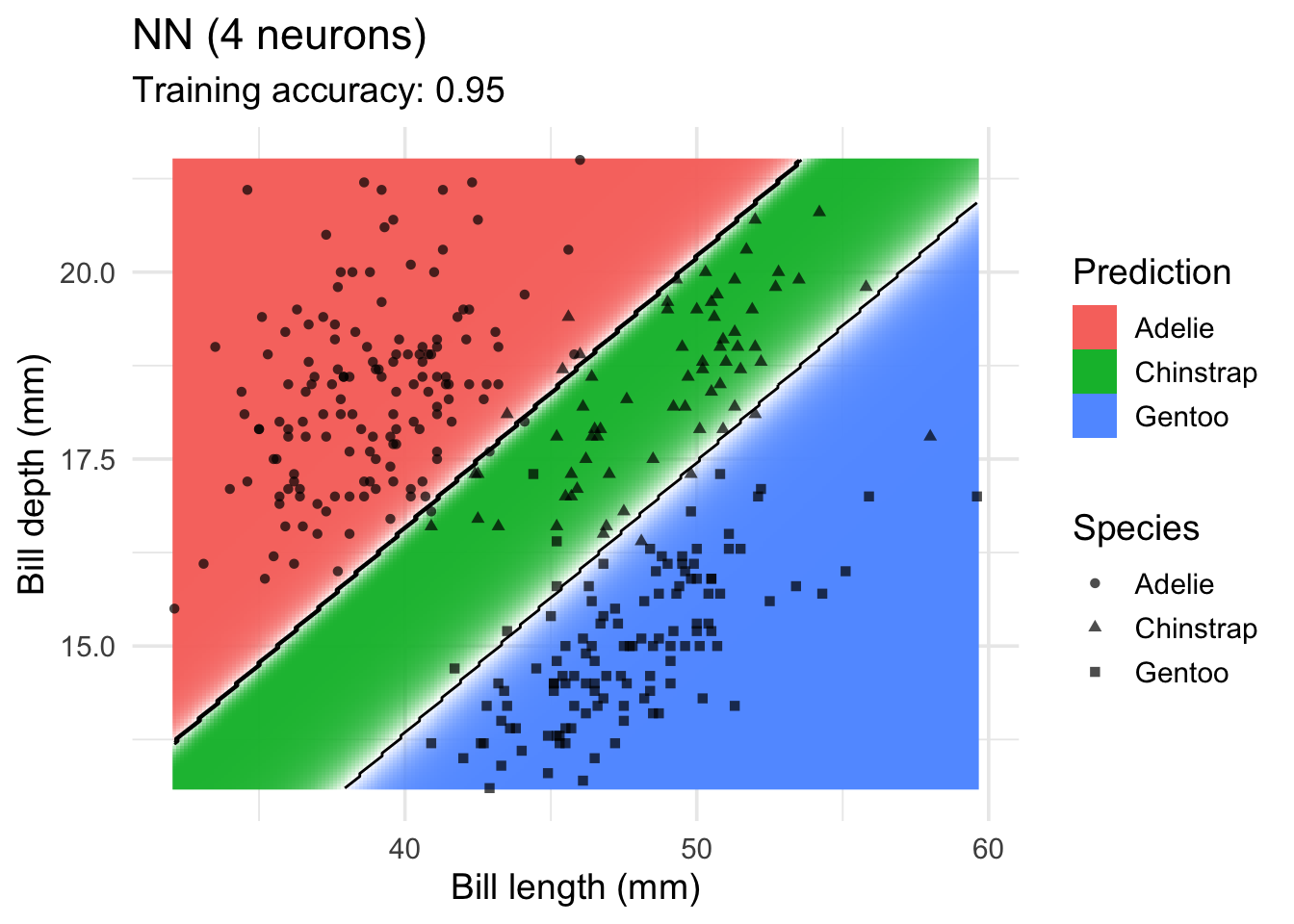
model <-x |> nnet::nnet(species ~ ., data = _, size = 10, trace = FALSE)
decisionplot(model, x, class_var = "species",
predict_type = c("class", "raw")) +
labs(title = "NN (10 neurons)",
x = "Bill length (mm)",
y = "Bill depth (mm)",
shape = "Species",
fill = "Prediction")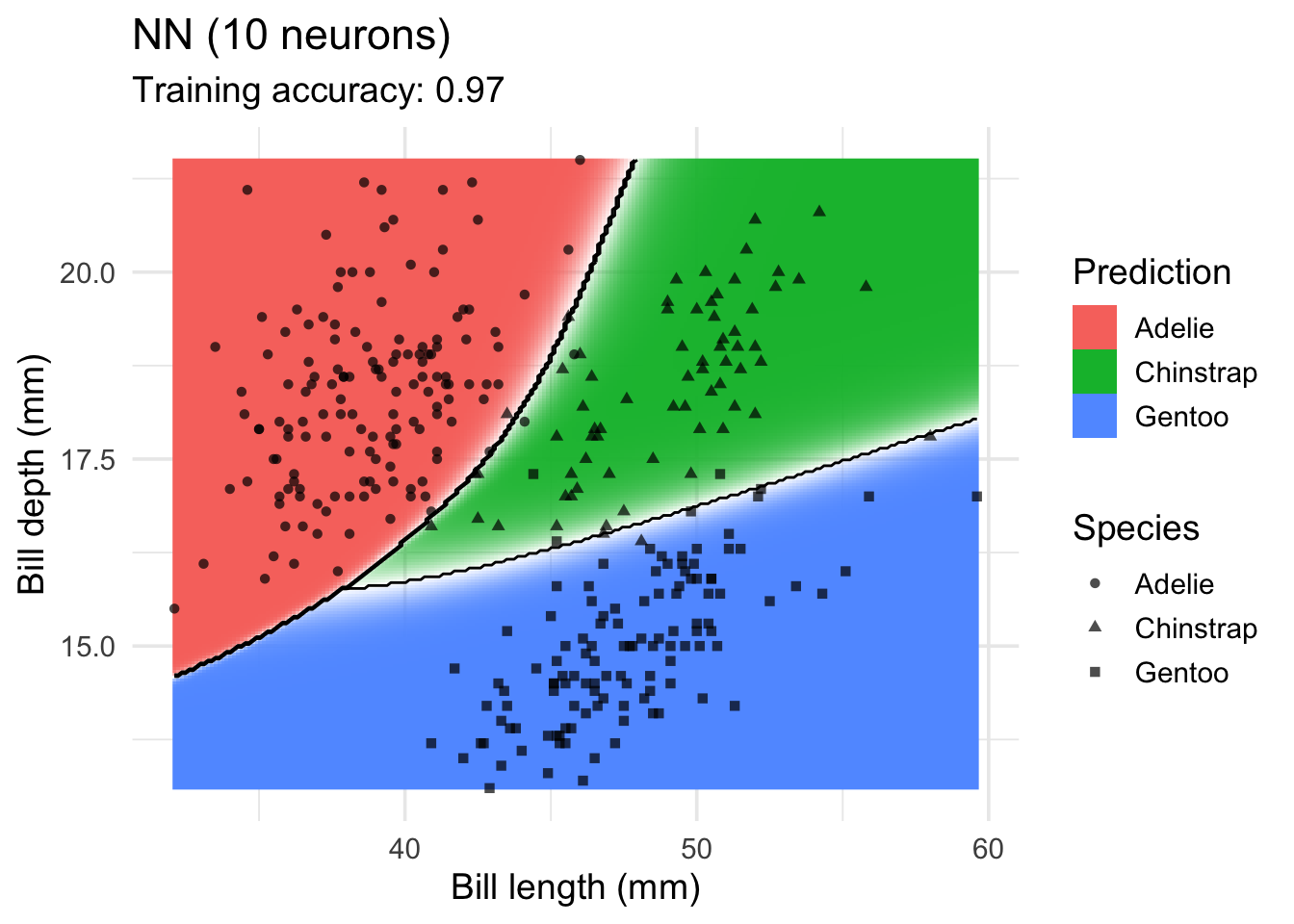
Circle Dataset
This set is not linearly separable!
set.seed(1000)
x <- mlbench::mlbench.circle(500)
###x <- mlbench::mlbench.cassini(500)
###x <- mlbench::mlbench.spirals(500, sd = .1)
###x <- mlbench::mlbench.smiley(500)
x <- cbind(as.data.frame(x$x), factor(x$classes))
colnames(x) <- c("x", "y", "class")
x <- as_tibble(x)
x# A tibble: 500 × 3
x y class
<dbl> <dbl> <fct>
1 -0.344 0.448 1
2 0.518 0.915 2
3 -0.772 -0.0913 1
4 0.382 0.412 1
5 0.0328 0.438 1
6 -0.865 -0.354 2
7 0.477 0.640 2
8 0.167 -0.809 2
9 -0.568 -0.281 1
10 -0.488 0.638 2
# ℹ 490 more rowsggplot(x, aes(x = x, y = y, color = class)) +
geom_point() +
theme_minimal(base_size = 14)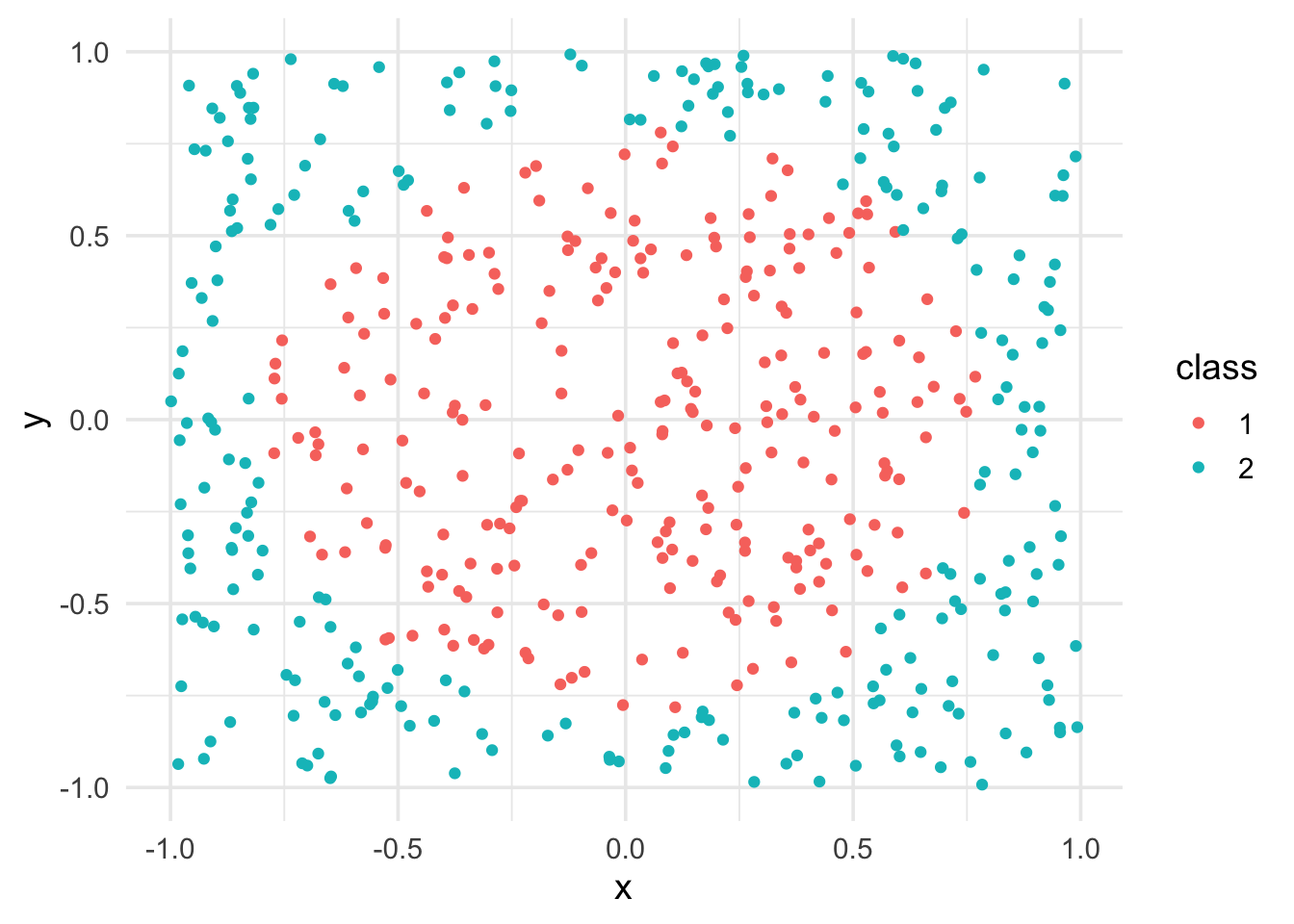
K-Nearest Neighbors Classifier
model <- x |> caret::knn3(class ~ ., data = _, k = 1)
decisionplot(model, x, class_var = "class") +
labs(title = "kNN (1 neighbor)",
shape = "Class",
fill = "Prediction")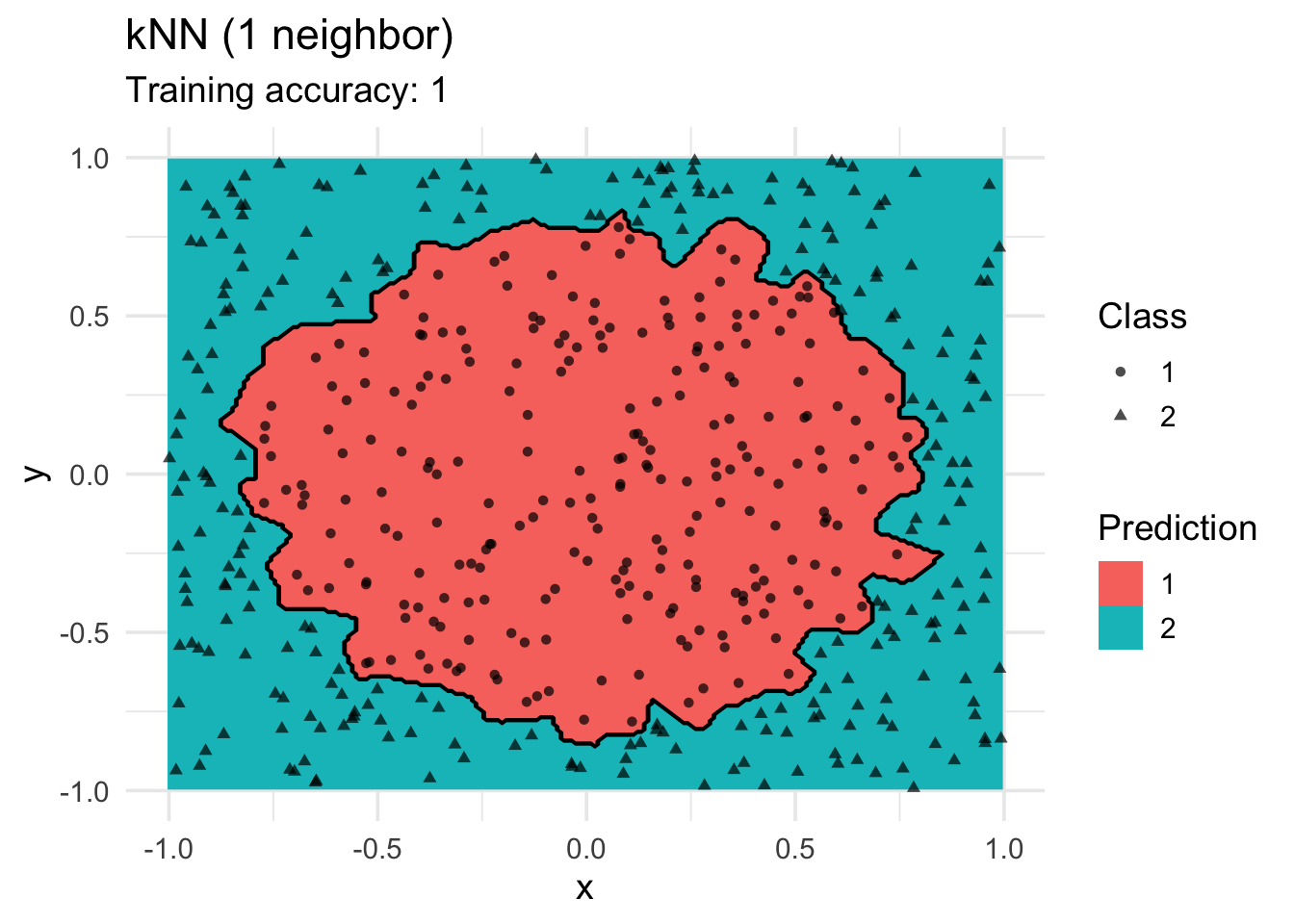
model <- x |> caret::knn3(class ~ ., data = _, k = 10)
decisionplot(model, x, class_var = "class") +
labs(title = "kNN (10 neighbor)",
shape = "Class",
fill = "Prediction")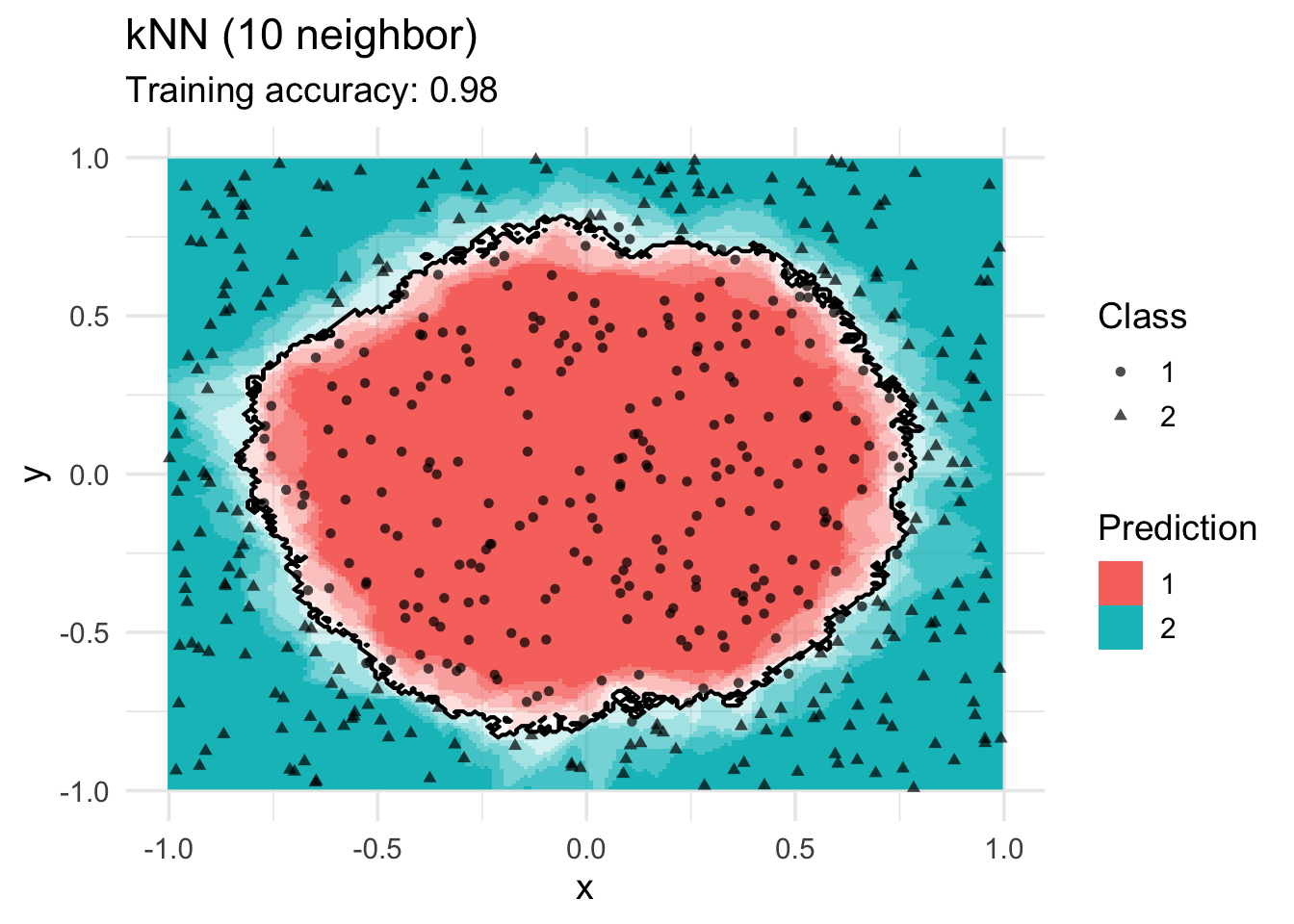
Naive Bayes Classifier
model <- x |> e1071::naiveBayes(class ~ ., data = _)
decisionplot(model, x, class_var = "class",
predict_type = c("class", "raw")) +
labs(title = "naive Bayes",
shape = "Class",
fill = "Prediction")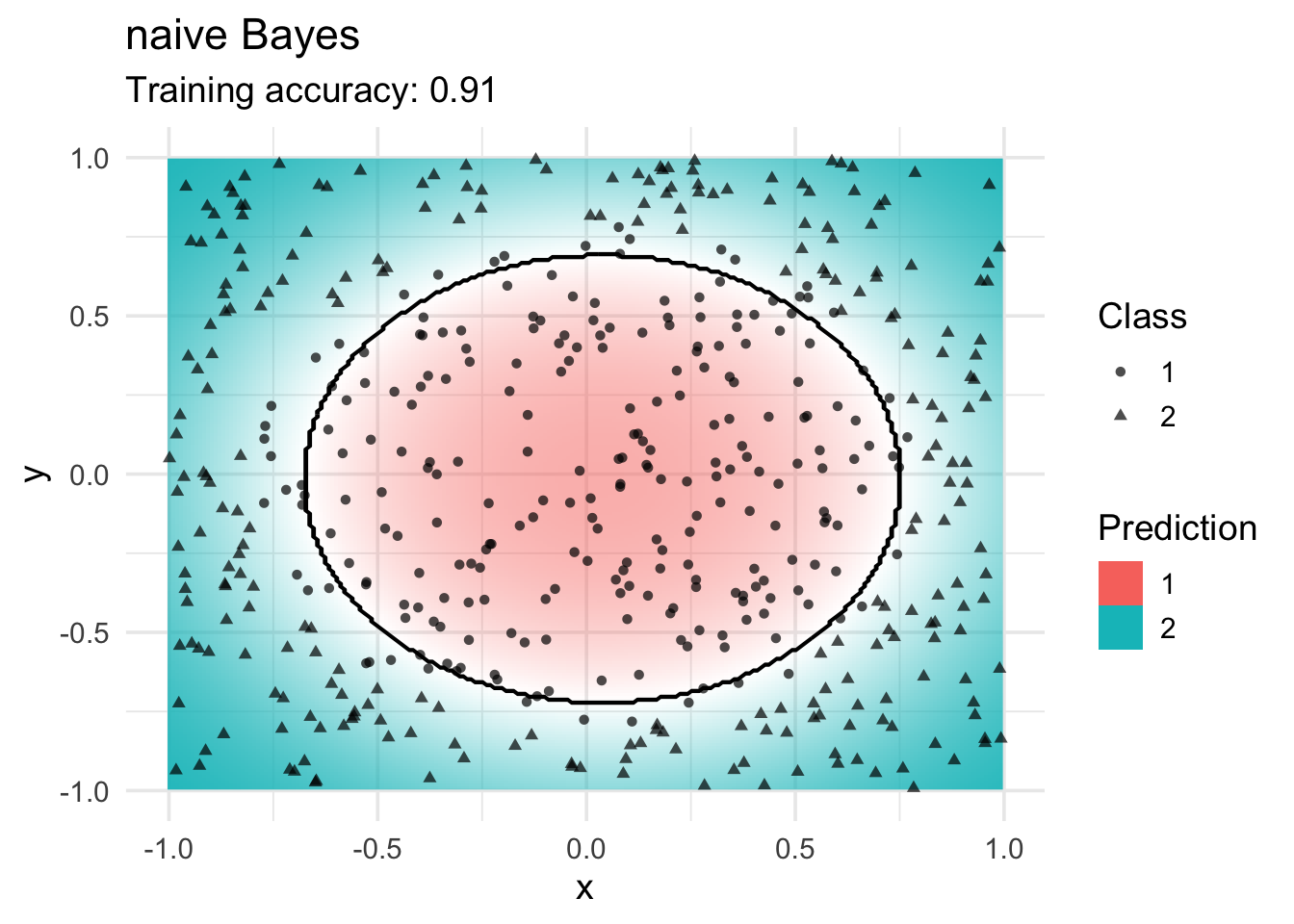
Linear Discriminant Analysis
LDA cannot find a good model since the true decision boundary is not linear.
model <- x |> MASS::lda(class ~ ., data = _)
decisionplot(model, x, class_var = "class") +
labs(title = "LDA",
shape = "Class",
fill = "Prediction")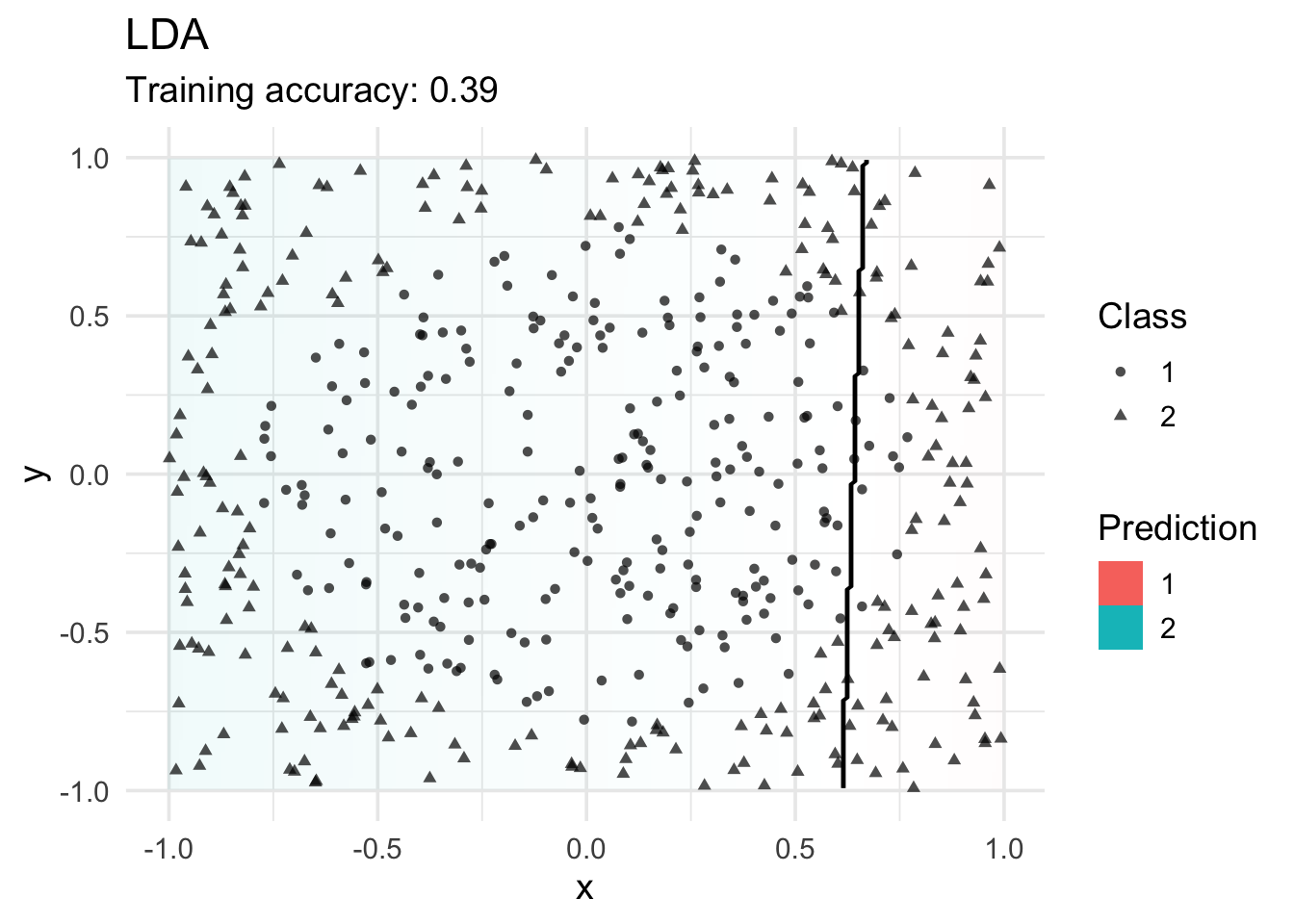
Logistic Regression (implemented in nnet)
Multinomial logistic regression is an extension of logistic regression to problems with more than two classes. It also tries to find a linear decision boundary.
model <- x |> nnet::multinom(class ~., data = _)# weights: 4 (3 variable)
initial value 346.573590
final value 346.308371
convergeddecisionplot(model, x, class_var = "class") +
labs(title = "Multinomial Logistic Regression",
shape = "Class",
fill = "Prediction")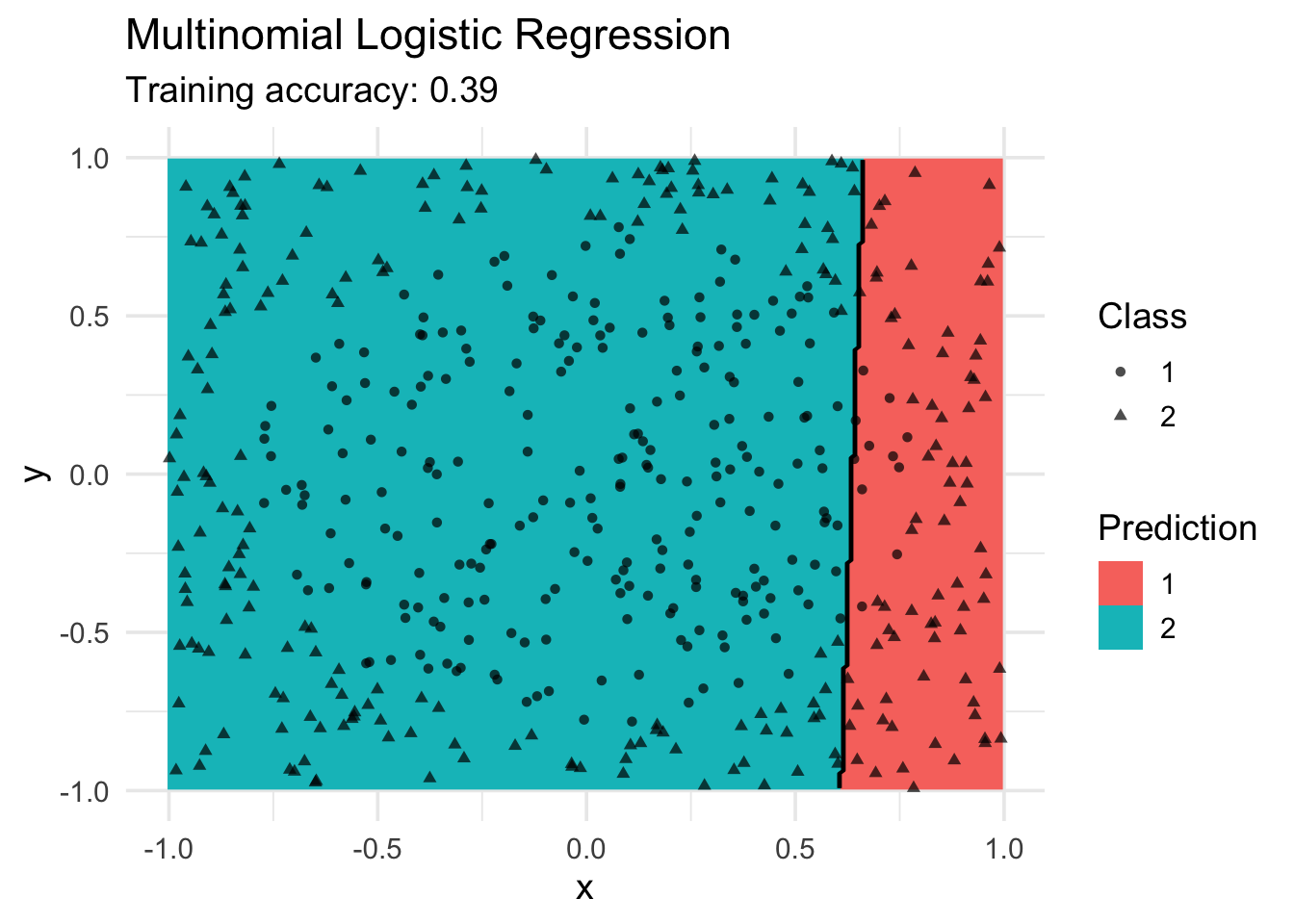
Decision Trees
model <- x |> rpart::rpart(class ~ ., data = _)
decisionplot(model, x, class_var = "class") +
labs(title = "CART",
shape = "Class",
fill = "Prediction")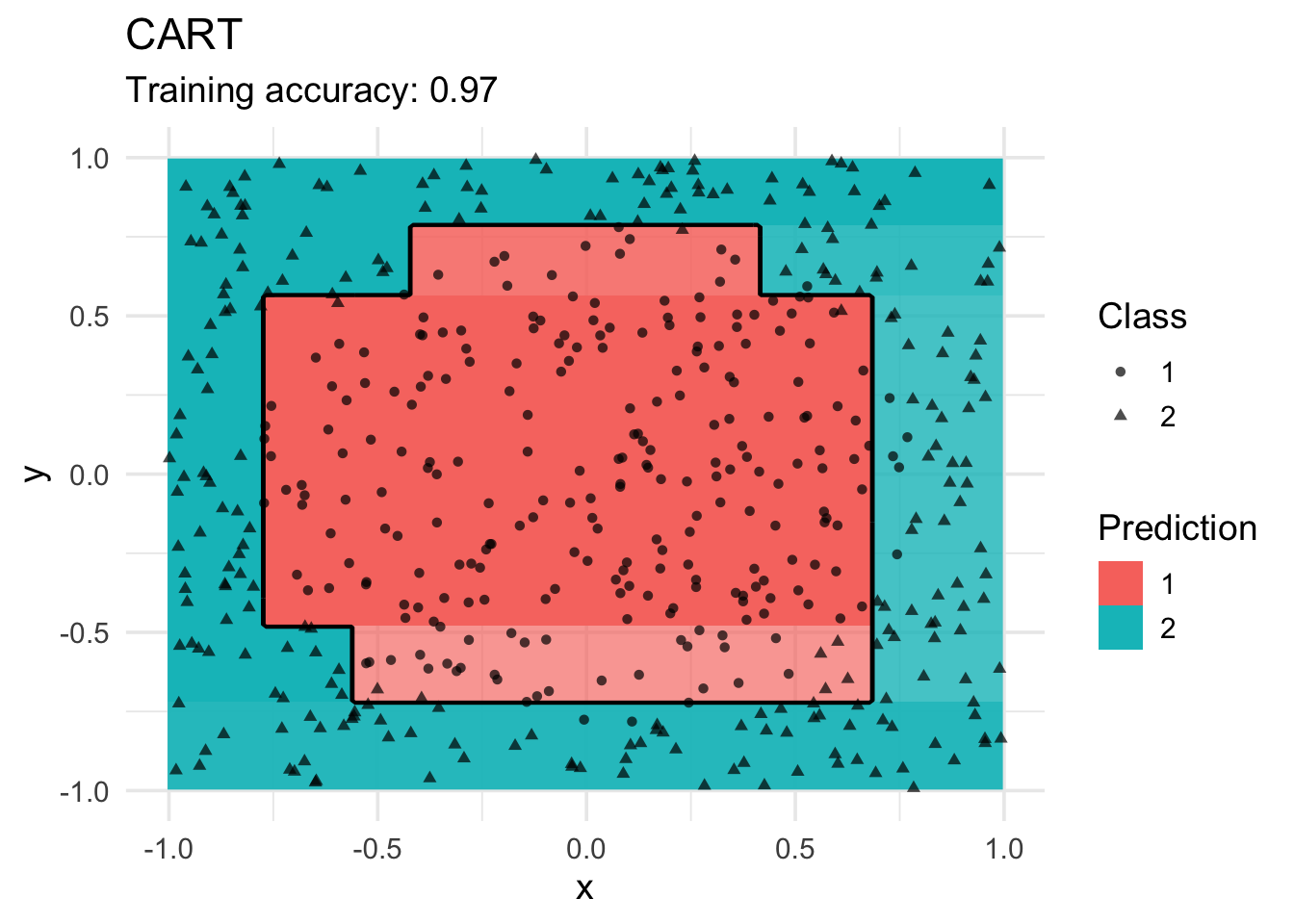
model <- x |> rpart::rpart(class ~ ., data = _,
control = rpart.control(cp = 0.001, minsplit = 1))
decisionplot(model, x, class_var = "class") +
labs(title = "CART (overfitting)",
shape = "Class",
fill = "Prediction")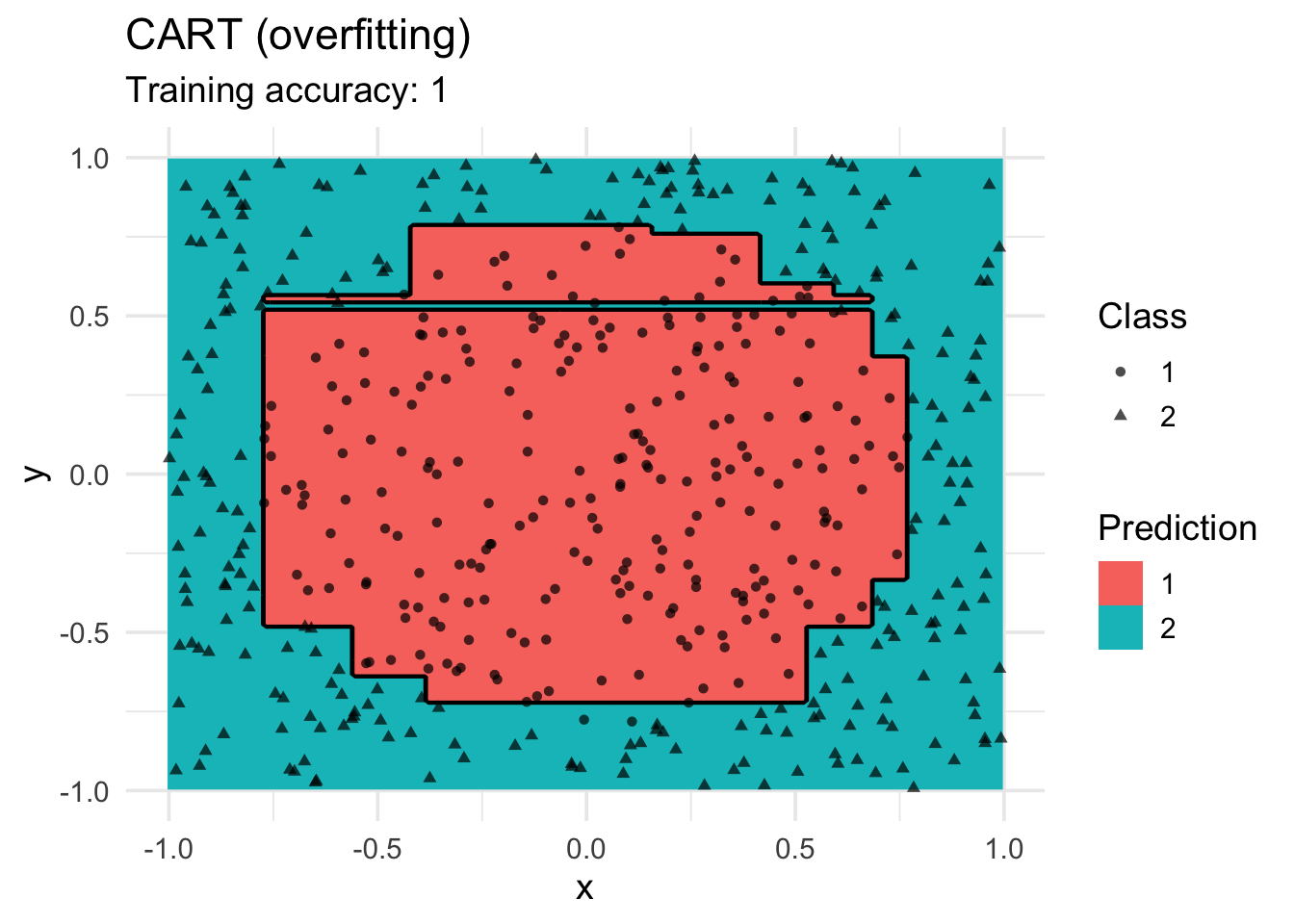
model <- x |> C50::C5.0(class ~ ., data = _)
decisionplot(model, x, class_var = "class") +
labs(title = "C5.0",
shape = "Class",
fill = "Prediction")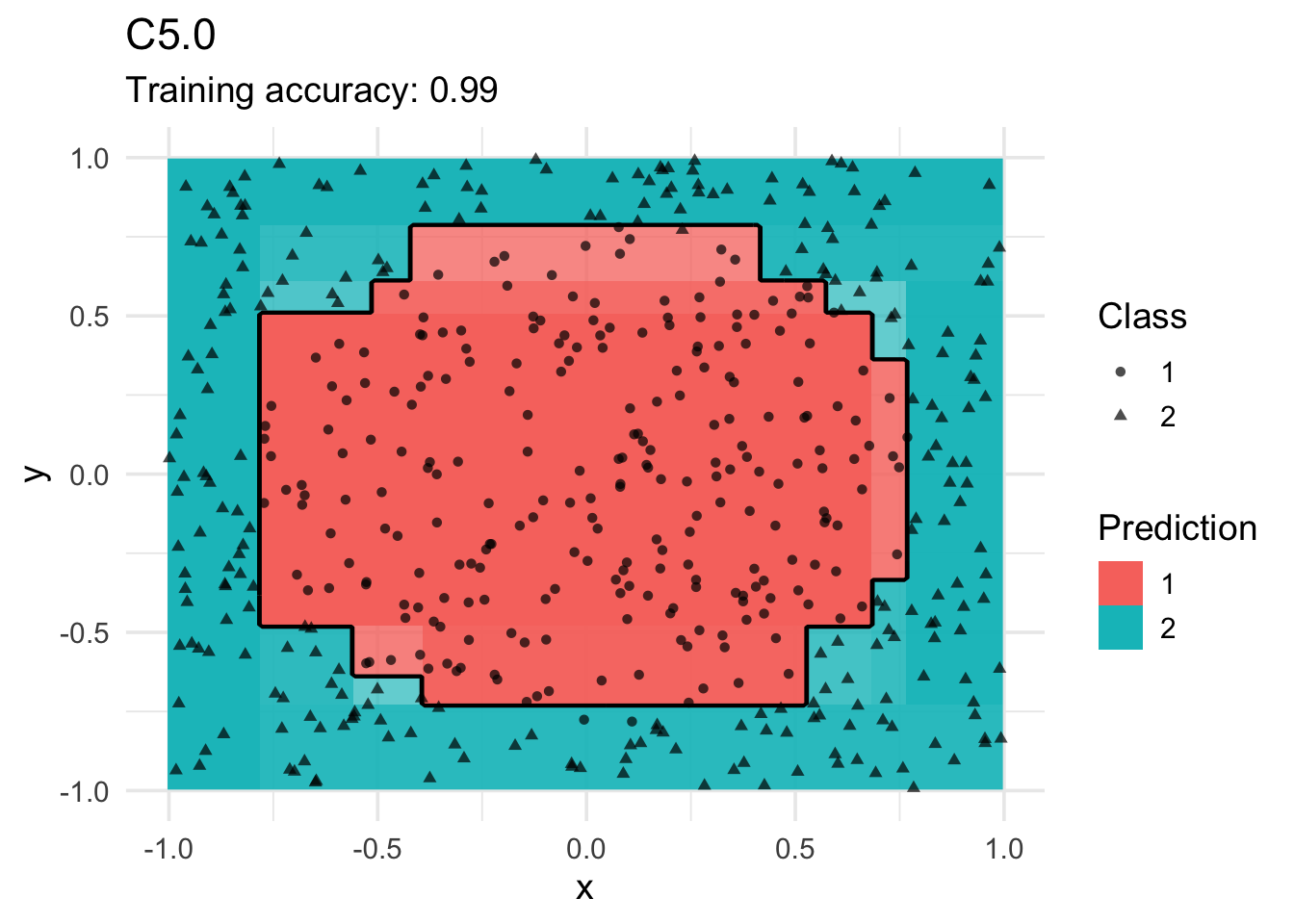
library(randomForest)
model <- x |> randomForest(class ~ ., data = _)
decisionplot(model, x, class_var = "class") +
labs(title = "Random Forest",
shape = "Class",
fill = "Prediction")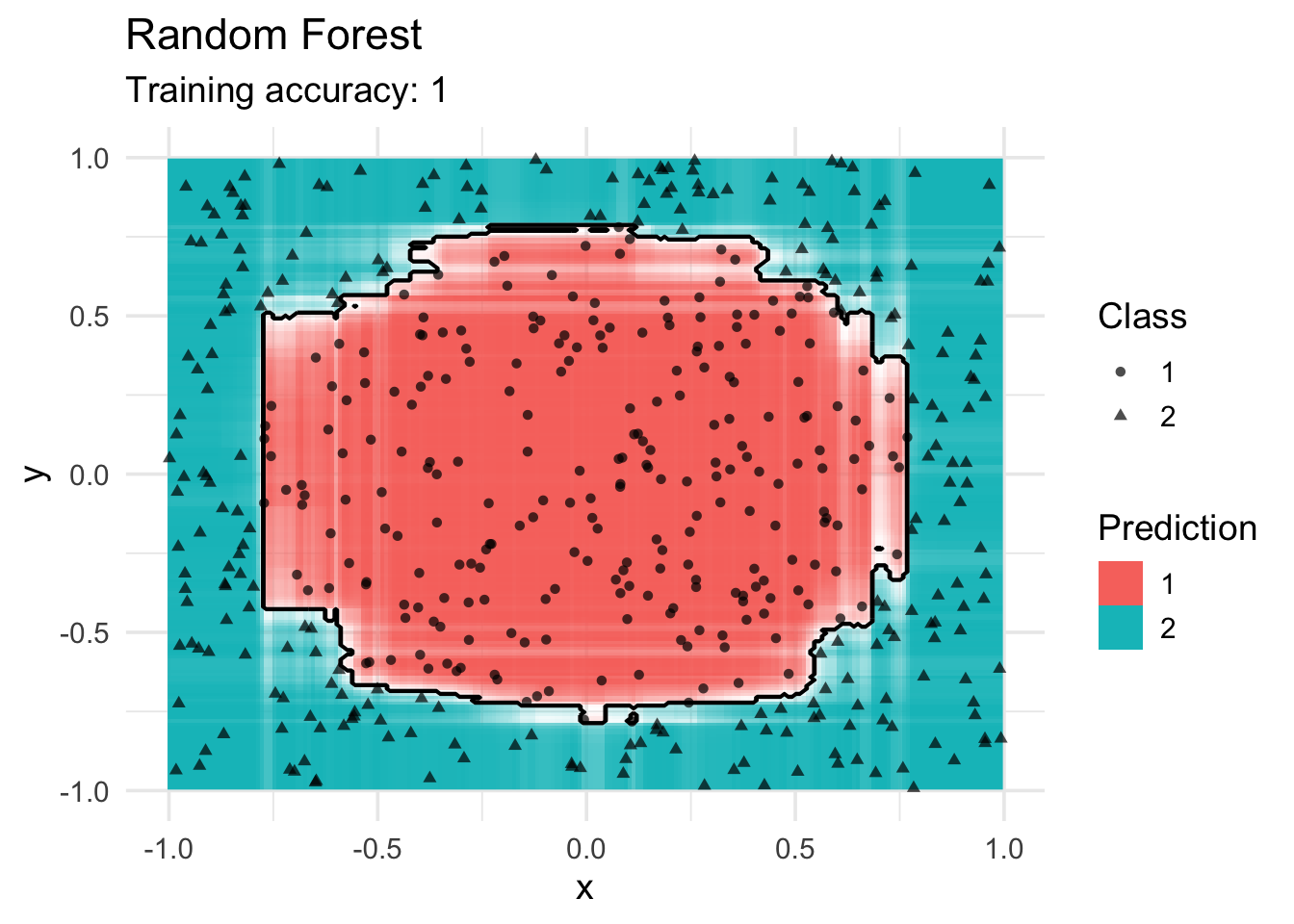
SVM
Linear SVM does not work for this data.
model <- x |> e1071::svm(class ~ ., data = _, kernel = "linear")
decisionplot(model, x, class_var = "class") +
labs(title = "SVM (linear kernel)",
shape = "Class",
fill = "Prediction")Warning: Computation failed in `stat_contour()`
Caused by error in `if (zero_range(range)) ...`:
! missing value where TRUE/FALSE needed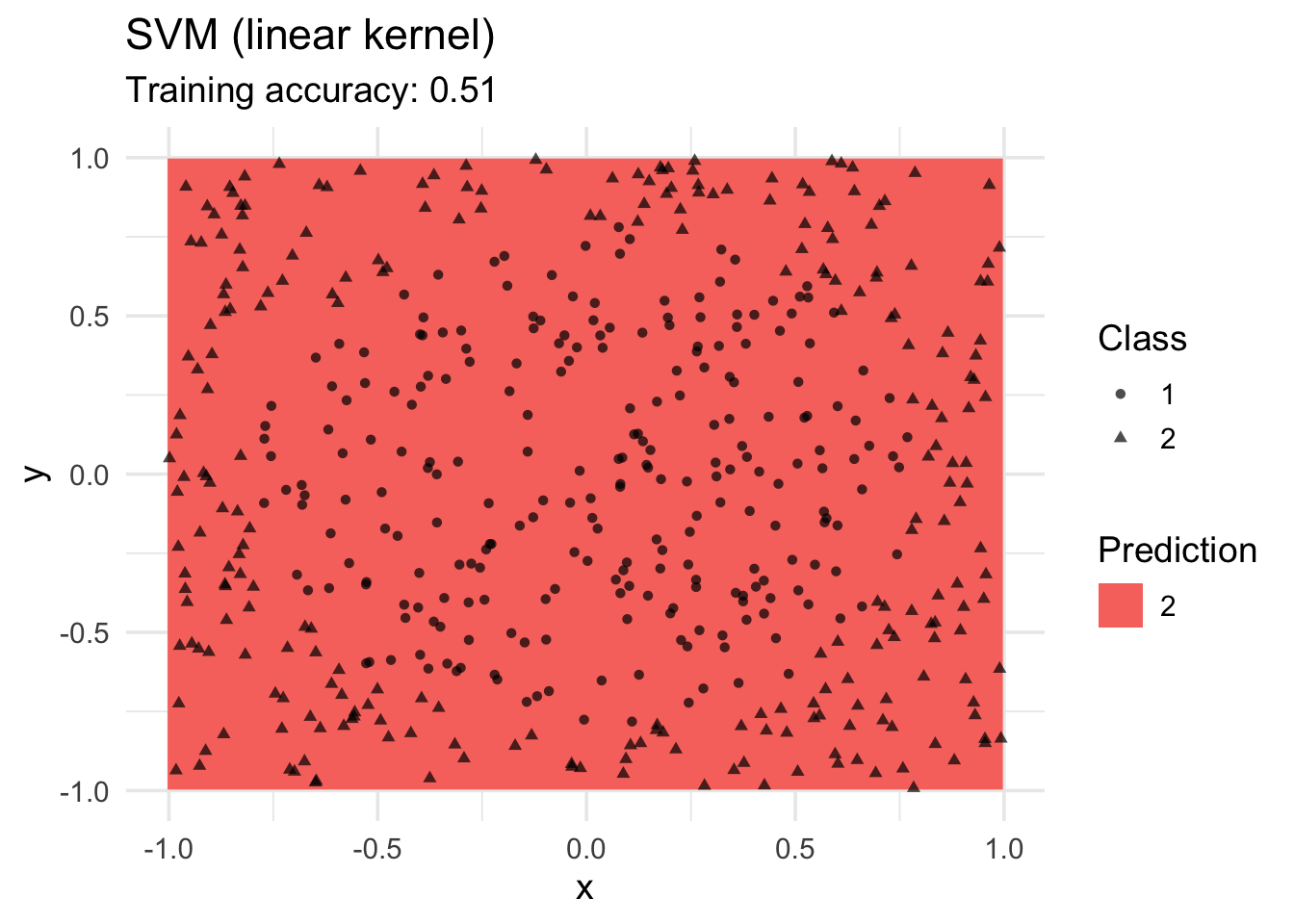
model <- x |> e1071::svm(class ~ ., data = _, kernel = "radial")
decisionplot(model, x, class_var = "class") +
labs(title = "SVM (radial kernel)",
shape = "Class",
fill = "Prediction")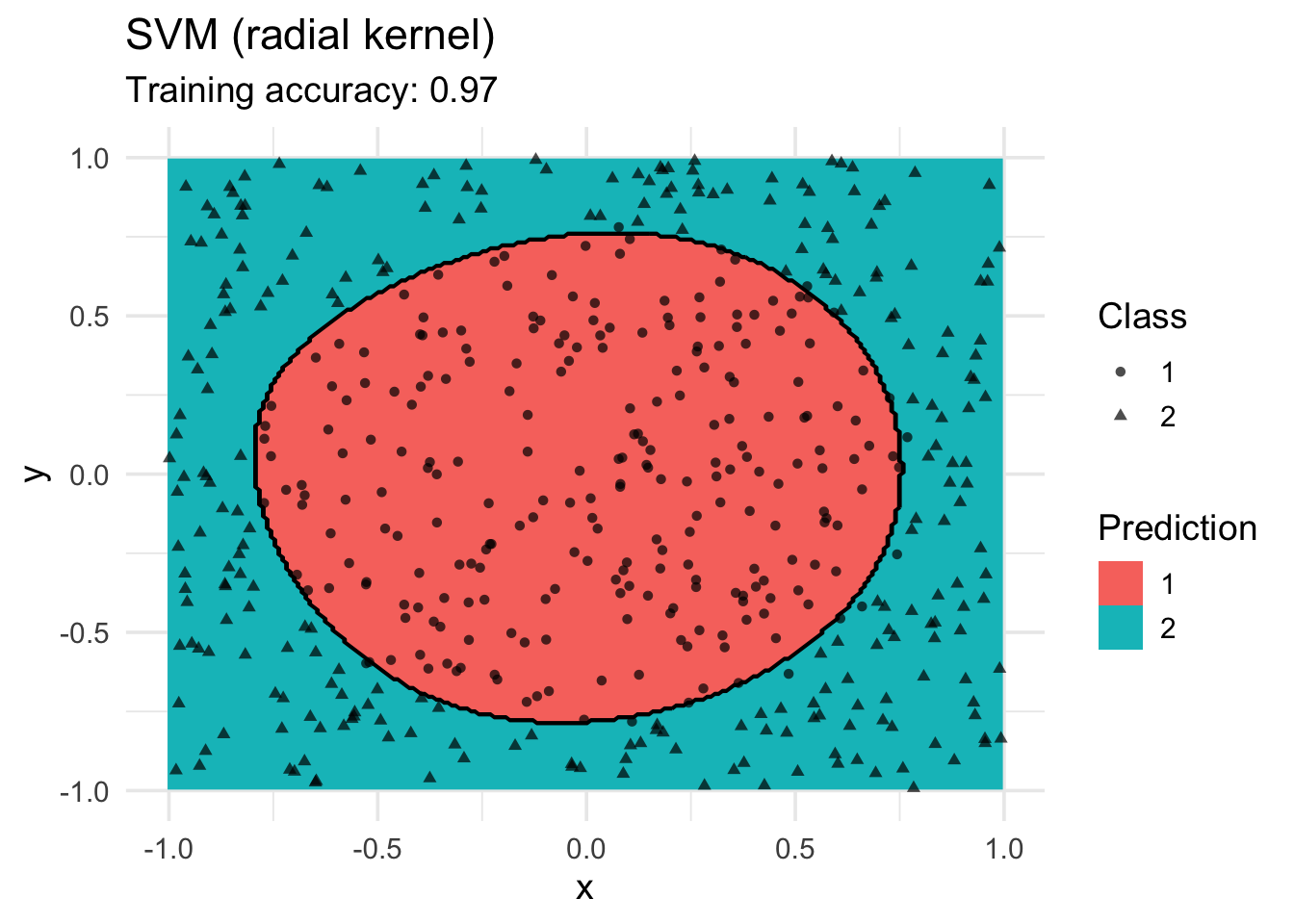
model <- x |> e1071::svm(class ~ ., data = _, kernel = "polynomial")
decisionplot(model, x, class_var = "class") +
labs(title = "SVM (polynomial kernel)",
shape = "Class",
fill = "Prediction")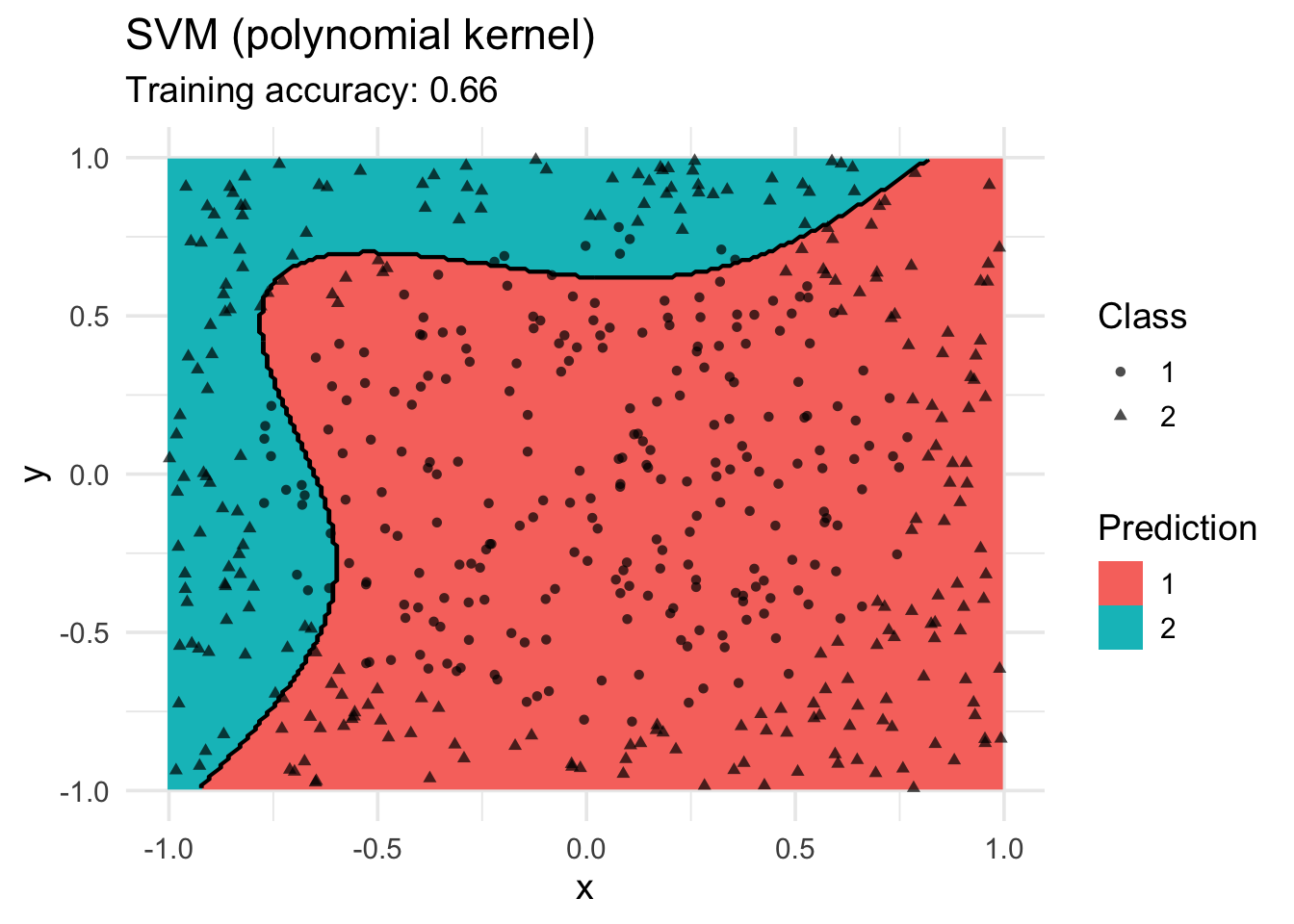
model <- x |> e1071::svm(class ~ ., data = _, kernel = "sigmoid")
decisionplot(model, x, class_var = "class") +
labs(title = "SVM (sigmoid kernel)",
shape = "Class",
fill = "Prediction")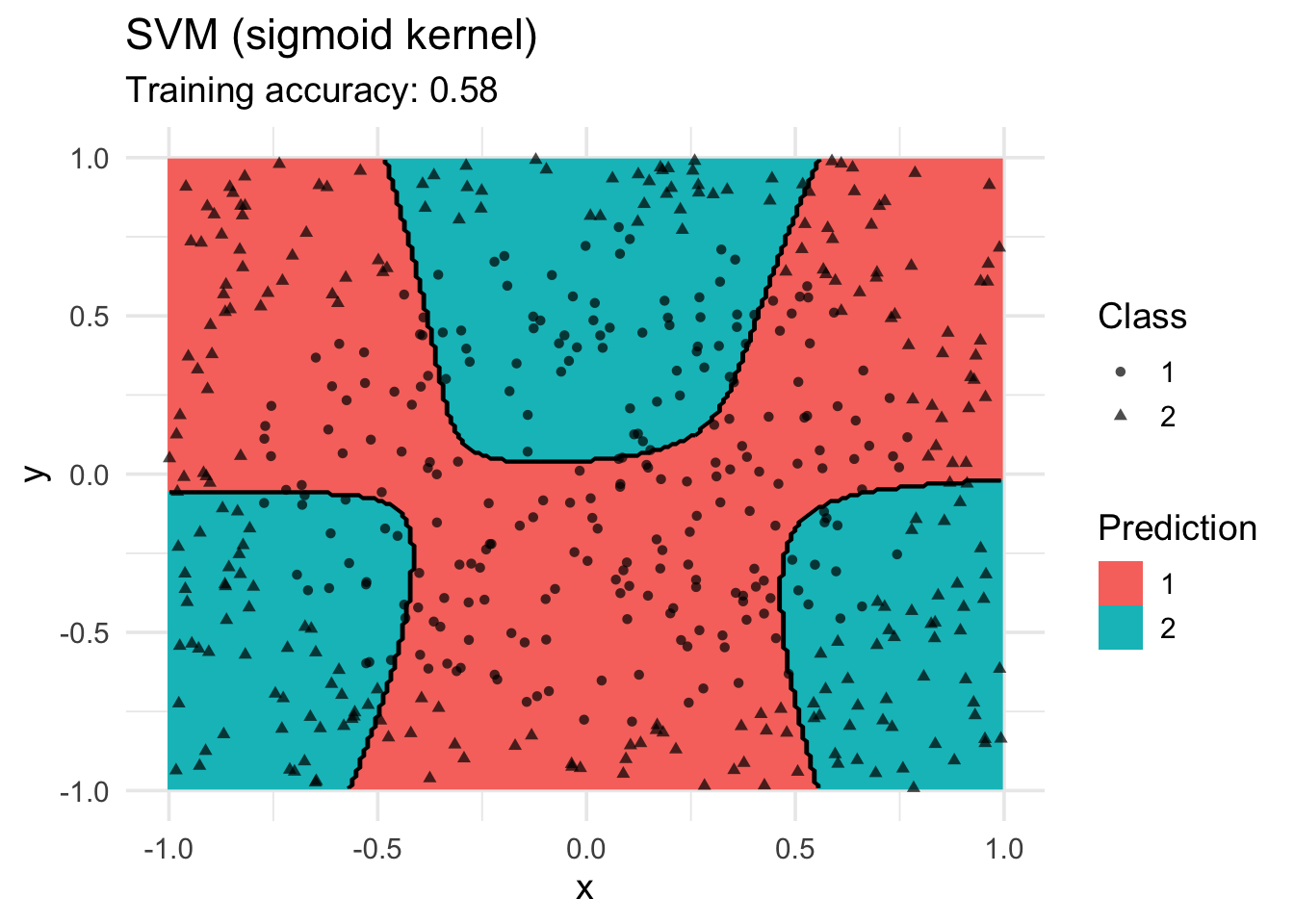
Single Layer Feed-forward Neural Networks
model <-x |> nnet::nnet(class ~ ., data = _, size = 1, trace = FALSE)
decisionplot(model, x, class_var = "class",
predict_type = c("class")) +
labs(title = "NN (1 neuron)",
shape = "Class",
fill = "Prediction")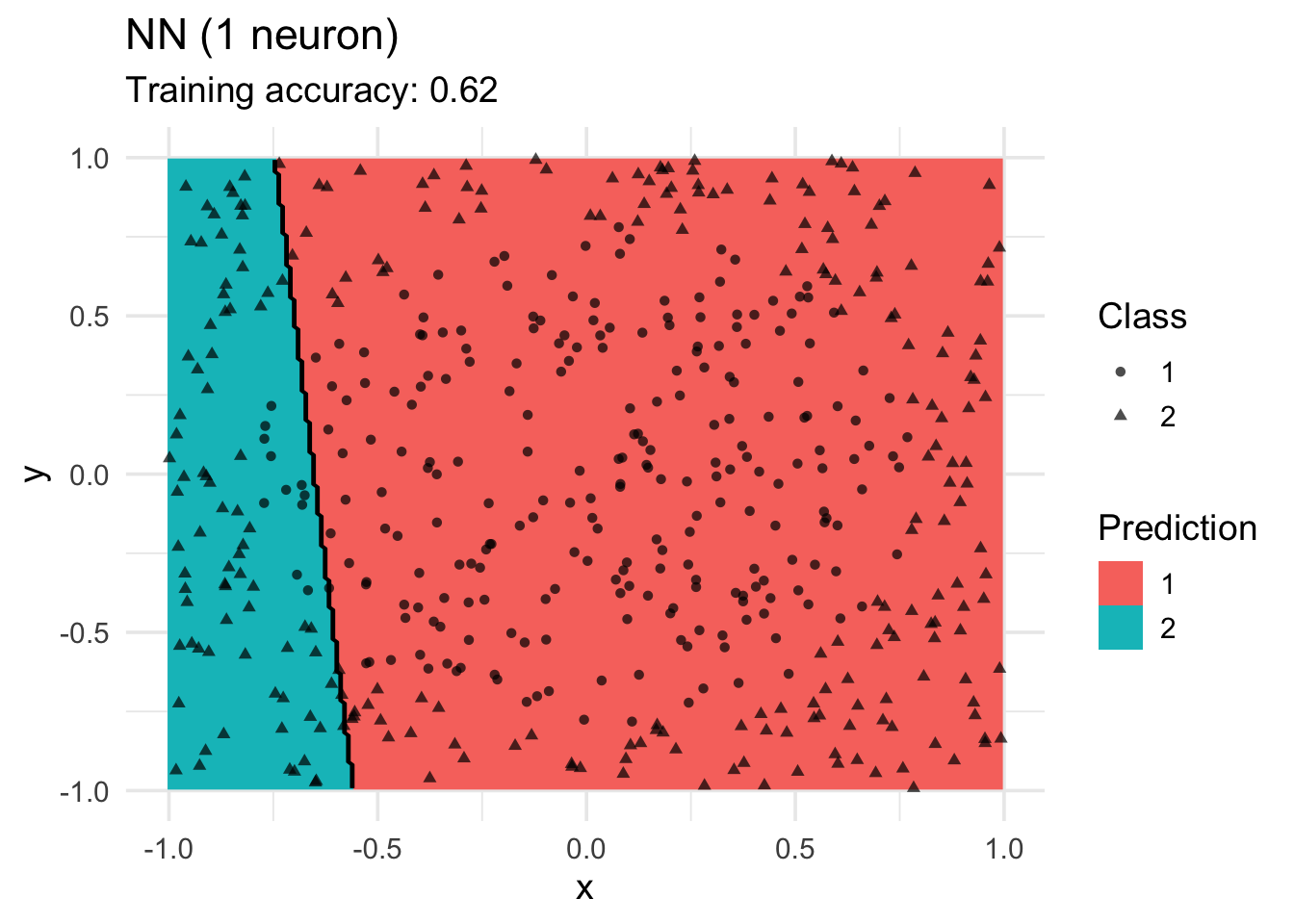
model <-x |> nnet::nnet(class ~ ., data = _, size = 2, trace = FALSE)
decisionplot(model, x, class_var = "class",
predict_type = c("class")) +
labs(title = "NN (2 neurons)",
shape = "Class",
fill = "Prediction")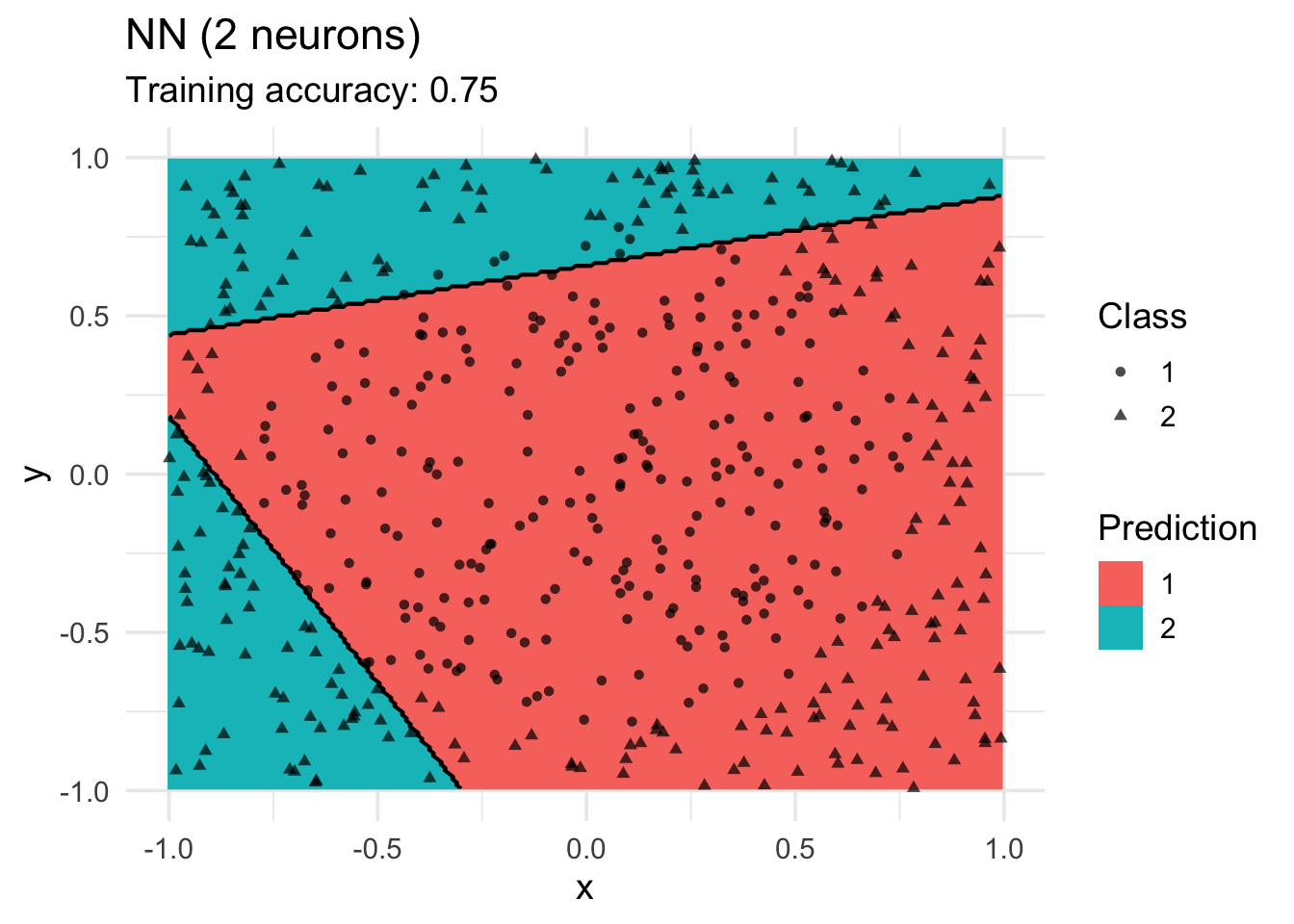
model <-x |> nnet::nnet(class ~ ., data = _, size = 4, trace = FALSE)
decisionplot(model, x, class_var = "class",
predict_type = c("class")) +
labs(title = "NN (4 neurons)",
shape = "Class",
fill = "Prediction")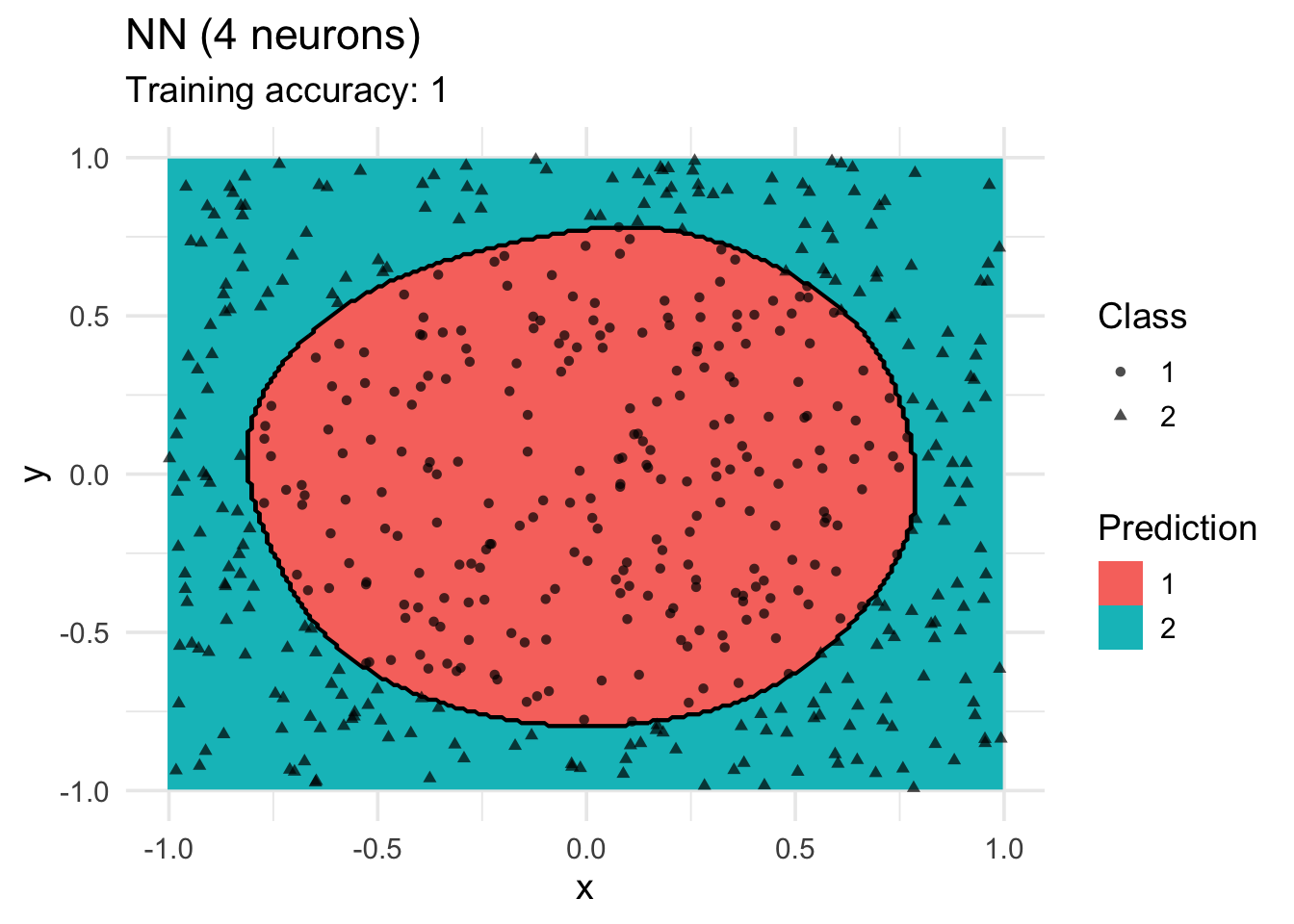
model <-x |> nnet::nnet(class ~ ., data = _, size = 10, trace = FALSE)
decisionplot(model, x, class_var = "class",
predict_type = c("class")) +
labs(title = "NN (10 neurons)",
shape = "Class",
fill = "Prediction")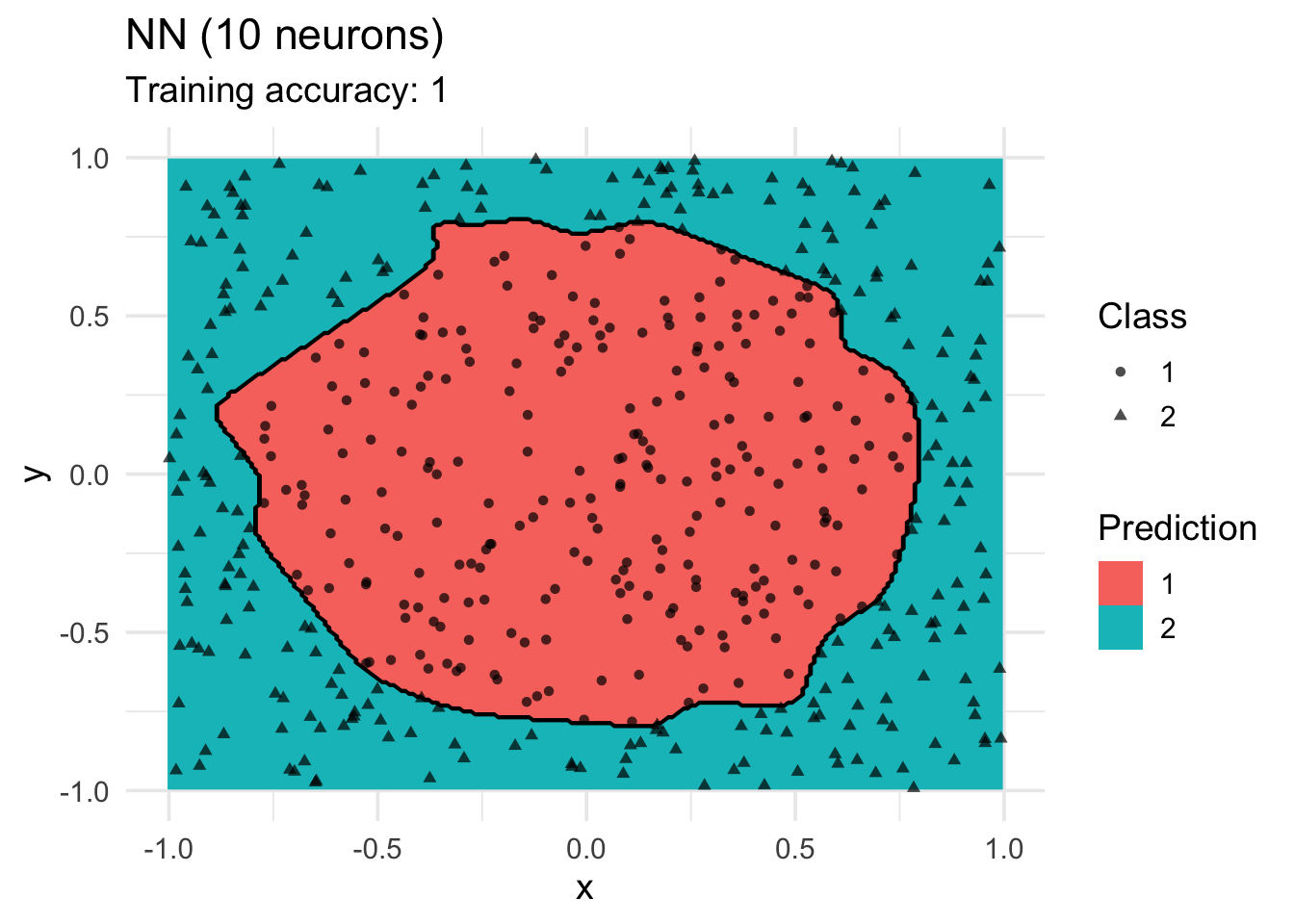
More Information on Classification with R
- Package caret: http://topepo.github.io/caret/index.html
- Tidymodels (machine learning with tidyverse): https://www.tidymodels.org/
- R taskview on machine learning: http://cran.r-project.org/web/views/MachineLearning.html
